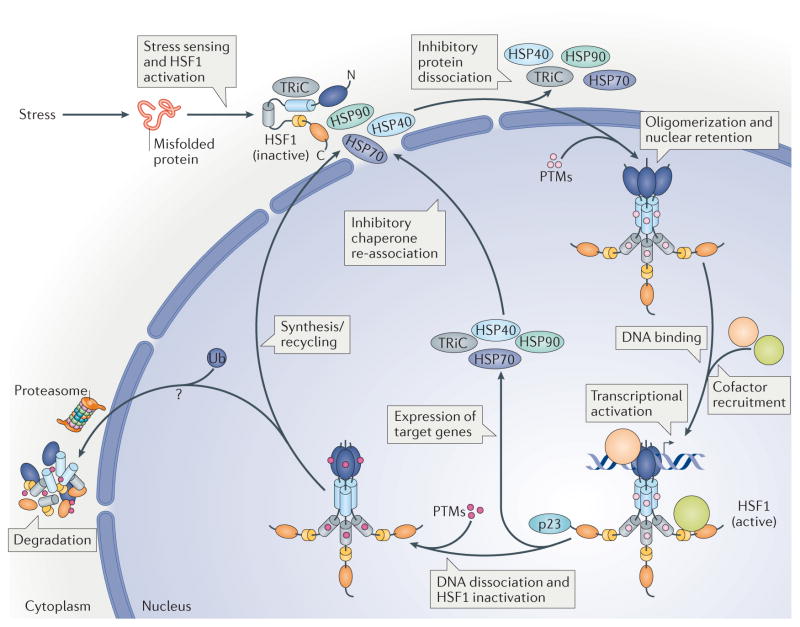
Figure 1. Heat shock transcription factor 1 activation cycle.
In response to proteotoxic stress conditions, heat shock transcription factor 1 (HSF1) is subject to a multistep activation and attenuation cycle. Inactive HSF1 monomers are retained in the cytoplasm in complex with regulatory proteins such as heat shock proteins (HSPs) 40, 70 and 90, as well as the cytosolic chaperonin TCP1 ring complex (TRiC). Upon stress sensing, HSF1 is activated, causing the dissociation of inhibitory proteins, HSF1 oligomerization and nuclear retention. HSF1 is modified by several activating post-translational modifications (PTMs) that promote DNA binding and transcriptional activation of target genes in concert with cofactor recruitment. HSF1 is then modified by different inhibitory PTMs, and by p23, causing DNA dissociation, HSF1 inactivation and HSF1 degradation (TABLE 1; see Supplementary information S3 (box)). It is currently unknown where HSF1 degradation occurs and to what extent HSF1 is newly synthesized or recycled into the cytoplasm. Ultimately, after attenuation, HSF1 is maintained in the cytoplasm by an inhibitory protein complex in a negative-feedback mechanism. Colour code: DNA-binding domain (dark blue), leucine zipper oligomerization domain LZ1 3 (light blue), regulatory domain (grey), LZ4 (yellow) and activation domain (orange).