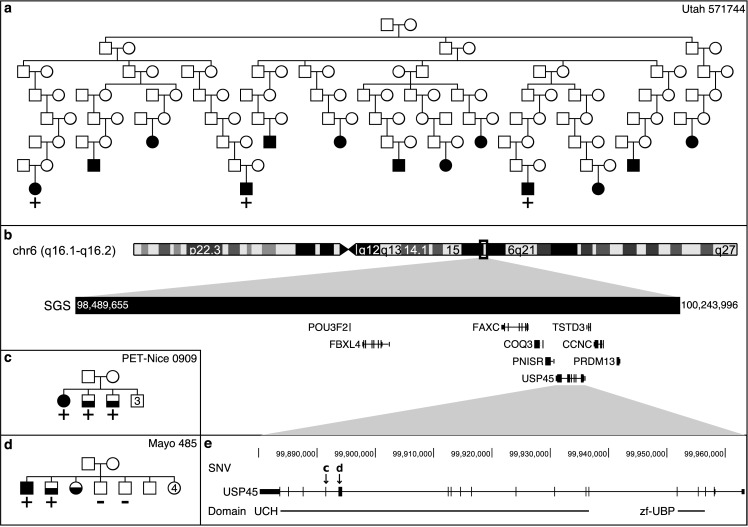
Fig 2. Significant SGS, pedigrees, and segregating SNVs.
In pedigrees, MM cases are fully shaded and MGUS cases are half shaded. Numbers indicate multiple individuals. a) Utah pedigree, 571744, sharing the genome-wide significant SGS. The pedigree is trimmed to allow for viewing (37 MM confirmed cases are known in this pedigree, 3 were ascertained and genotyped). + indicates the genotyped MM cases that are SGS carriers, − indicates genotyped and non-carriers, no carrier status indicates not genotyped. Note–the genealogy extends beyond SEER cancer registry data. MGUS status is unknown in this pedigree. b) Genomic region of significant SGS. c) INSERM pedigree carrying the stop gain SNV marked by “c” in box e. 1 MM and 2 MGUSs carry the SNV. d) Mayo Clinic pedigree carrying the missense SNV marked by “d” in box e. 1 MM and 1 MGUS carry the SNV, but 2 unaffected siblings do not carry the SNV. e) Risk candidate gene, USP45, has 2 segregating SNVs in the ubiquitin C-terminal hydrolase 2 (UCH) domain.