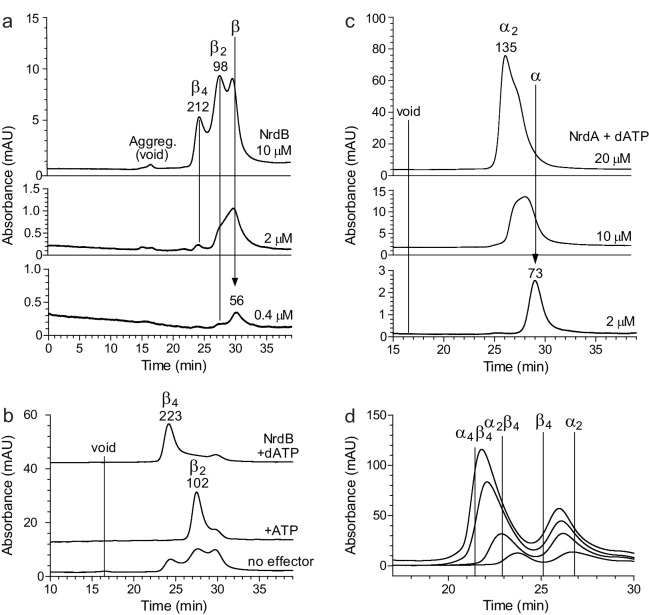
Figure 5. Size exclusion chromatography of L. blandensis RNR in the absence and presence of 3 mM ATP or 0.5 mM dATP.
Proteins at concentrations of 20 μM each were pre-incubated separately or mixed together for 10 min at room temperature in the absence (black dotted line) or presence of ATP (blue dashed line) or dATP (red solid line), centrifuged and applied to the column equilibrated with SEC buffer at 7°C. Panels show NrdB (a), NrdA (b), and an equimolar mixture of NrdA and NrdB (c). C Insert: SDS PAGE of the eluted proteins run in the presence and absence of the indicated effectors. Elution positions of fractions applied to gel are indicated by red (in the presence of dATP) and blue (in the presence of ATP) arrows, respectively. (d) Summary of multiple SEC experiments varying protein concentrations of NrdA (10–113 μM), NrdB (5–150 μM) and mixtures of NrdA and NrdB at ratios of 1:1 or 1:2. Molecular masses and standard deviations are calculated for 3–5 experiments, and closest estimated complex stoichiometry are shown in parenthesis, with major species underlined when appropriate (see Figure 5—figure supplement 1 for details).