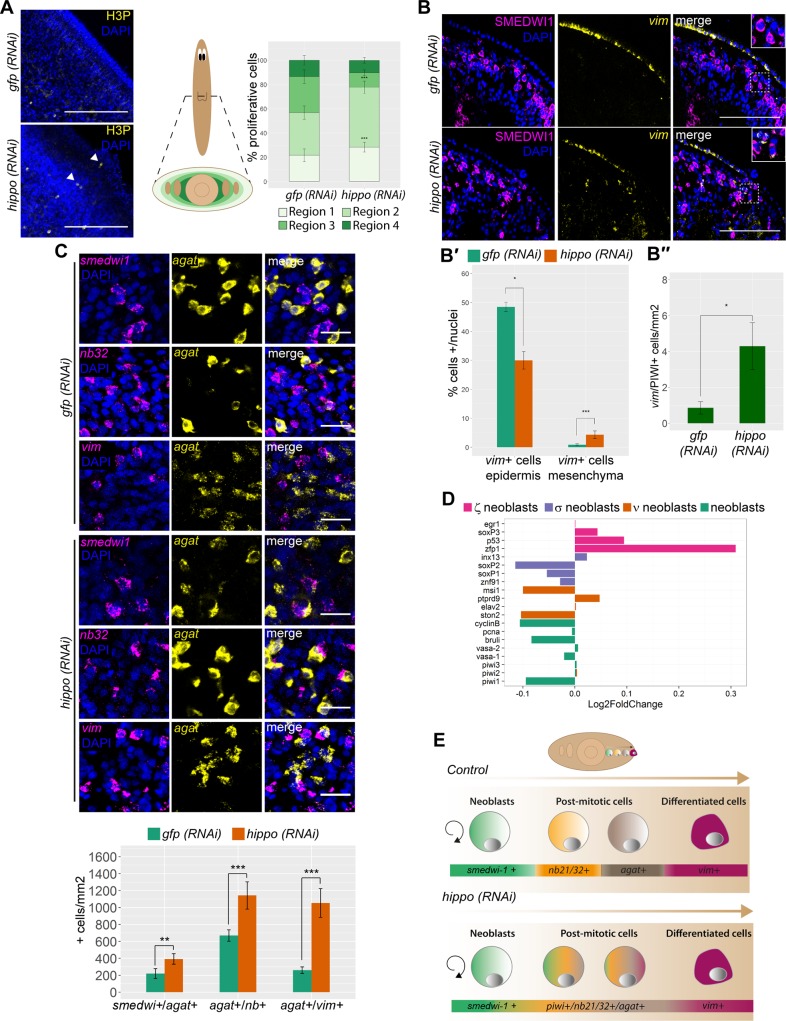
Fig 5. hippo defines the expression boundaries of epidermal markers.
(A) Immunostaining with anti-H3P in the subepidermal region of hippo (RNAi) animals and controls. Nuclei are labeled in blue. A schematic indicating the different regions quantified, from outermost (light green) to innermost (dark green), is shown, as well as corresponding quantification of the number of H3P+ cells in the different regions along the mediolateral axis (n ≥ 6). The total number of H3P+ cells found in each region was normalized to the selected area, as indicated in the schematic. White arrows indicate H3P+ cells. (B) FISH for vim combined with anti-SMEDWI-1 immunostaining in transverse sections from hippo (RNAi) animals and controls. (B′) Corresponding quantification of vim+ cells in the epidermis and mesenchyme; n ≥ 6. (B″) Corresponding quantification of vim+/SMEDWI-1+ cells (n ≥ 6). (C) Double FISH with markers of different epidermal cell progenitors: smedwi-1/agat, agat/nb32, and agat/vim and corresponding quantification (n ≥ 6). (D) Graph showing the relative expression levels of different neoblast markers in the RNAseq of hippo (RNAi) animals [78, 79]. Error bars represent standard deviation. Data were analyzed by Student t test. *p < 0.05; **p < 0.01; ***p < 0.001. (E) Schematic showing the consequences of hippo inhibition in the epidermal lineage. In wild-type animals, the onset of stem cell differentiation results in the loss of smedwi-1 expression and the acquisition of postmitotic genes in a sequential manner (nb32 → agat → vim) to reach the fully differentiated state in the epidermis (vim+ cells). In hippo (RNAi) animals, the spatiotemporal transition of postmitotic epidermal cells is lost: smedwi-1, nb32, agat, and vim are co-expressed in the same cells and the number of fully differentiated cells is decreased. All experiments were performed in planarians subjected to hippo RNAi for 3 weeks. Quantifications in (B) and (C) correspond to the posterior half of the body of each animal. Data used in the generation of this figure can be found in S1 Data. Scale bars: 150 μm (A); 50 μm (B); 25 μm (C). FISH, fluorescent in situ hybridization; gfp, green fluorescent protein; H3P, phospho-histone-H3-Ser10; RNAi, RNA interference; vim, vimentin.