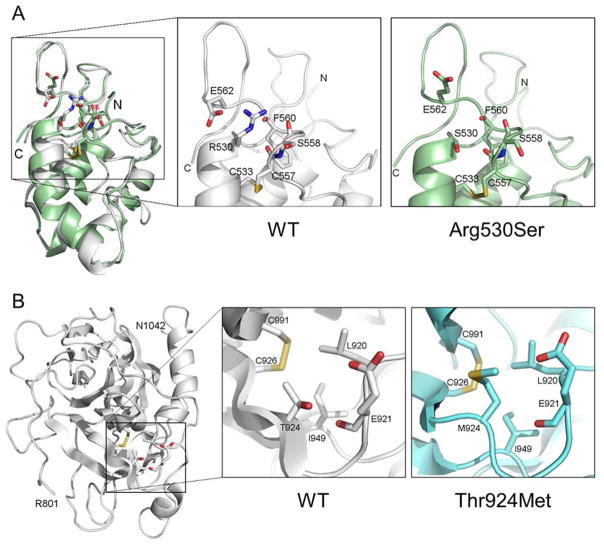
FIGURE 4.
Molecular models of corin Fz2 and protease domains. (A) Ribbon model of the corin Fz2 domain was based on the human frizzled 4 crystal structure. The model indicates that the R530 side chain makes a salt bridge with E562 and hydrogen bonds with the surrounding backbone oxygen atoms (left small box and enlarged middle box labeled with WT). The R530S substitution disrupts these hydrophilic interactions (enlarged right box labeled with Arg530Ser). (B) Ribbon model of the corin protease domain was based on the human plasminogen protease domain crystal structure. The model indicates that T924 sidechain oxygen atom forms hydrogen bonds with the E921 side chain and backbone oxygen atoms. The T924 methyl group also contributes to the hydrophobic interactions with C926, L920, and I949 (left small box and enlarged middle box labeled with WT). The substitution of hydrophilic T924 by a long-chain hydrophobic Met disrupts the hydrogen bond and the hydrophobic networks (enlarged right box labeled with Thr924Met). The corin activation site R801 is at the N-terminal of the protease domain