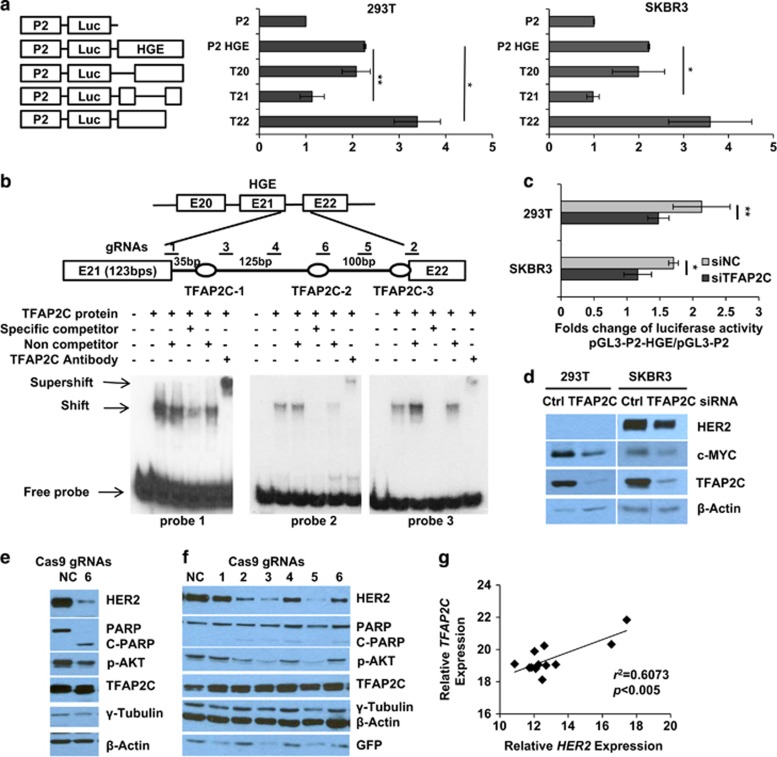
Figure 3.
TFAP2C positively regulates enhancer activity of the HGE. (a) Relative luciferase activities of reporter constructs bearing deletions in HGE region shown at left (T20: deletion of exon 20 and intron 20 in the pGL3-P2-HGE vector) in 293T and SKBR3 cells. Relative luciferase activities of the indicated constructs normalized to Renilla luciferase expression in cells co-transfected with pRL-TK-Rluc. (b) Schematic illustration of positions of three potential TFAP2C-binding sites corresponding to three probes for electrophoretic mobility shift assay (EMSA). Unlabeled 25-bp oligonucleotides or nonspecific competitor were added as indicated. + and − indicate the presence or absence of reaction components. Shifts, supershifts and free probes are indicated with arrows. (c) Fold change of relative luciferase activity of pGL3-P2-HGE normalized to that of pGL3-P2 vectors in 293T or SKBR3 cells transiently transfected with control (Ctrl) or TFAP2C siRNAs. (d) Expression of the indicated proteins as assessed by western blotting. Mean±s.d. in a and c were obtained from three independent experiments. Student's t-test was used to calculate the significance. *P<0.05, **P<0.001. (e, f). Six gRNAs were designed to introduce mutations in intron 21 as indicated in b. Virus infected (e) and transient transfection for 48 h (f) of CRISPR-Cas9-GFP vector with corresponding gRNAs in SKBR3 cells were assessed by western blotting. (g) RT-PCR used to determine a correlation between HER2 expression and expression of TFAP2C in panel of primary breast cancer samples (n=12). The correlation (r2) and significance (P-value) are shown.