Abstract
The gene encoding the ATPase of the chromatin remodeling SWI/SNF complexes SMARCA4 (BRG1) is often mutated or silenced in tumors, suggesting a role as tumor suppressor. Nonetheless, recent reports show requirement of SMARCA4 for tumor cells growth. Here, we performed a computational meta-analysis using gene expression, prognosis, and clinicopathological data to clarify the role of SMARCA4 and the alternative SWI/SNF ATPase SMARCA2 (BRM) in cancer. We show that while the SMARCA4 gene is mostly overexpressed in tumors, SMARCA2 is almost invariably downexpressed in tumors. High SMARCA4 expression was associated with poor prognosis in many types of tumors, including liver hepatocellular carcinoma (LIHC), and kidney renal clear cell carcinoma (KIRC). In contrast, high SMARCA2 expression was associated with good prognosis. We compared tumors with high versus low expression of SMARCA4 or SMARCA2 in LIHC and KIRC cohorts from The Cancer Genome Atlas. While a high expression of SMARCA4 is associated with aggressive tumors, a high expression of SMARCA2 is associated with benign differentiated tumors, suggesting that SMARCA4 and SMARCA2 play opposite roles in cancer. Our results demonstrate that expression of SMARCA4 and SMARCA2 have a high prognostic value and challenge the broadly accepted general role of SMARCA4 as a tumor suppressor.
Introduction
ATP-dependent chromatin remodeling is essential for almost every aspect of DNA metabolism including transcription, recombination, DNA repair, and DNA replication1,2. Therefore, it is not surprising that chromatin remodeling enzymes play a fundamental role in the development of cancer3,4. The first chromatin remodeling machinery identified was the SWI/SNF complex (also called the BAF complex), comprised in mammals by 11–15 subunits5–7. In fact, there is not a single SWI/SNF complex but rather a polymorphic family of complexes that also includes different members of small gene families. The enzymatic motor of the complexes are two mutually exclusive ATPases of the SNF2 family called SMARCA2 (also called BRAHMA, BRM)8 and SMARCA4 (also called BRAHMA RELATED GENE 1, BRG1)9. The mammalian SWI/SNF complexes have been involved in chromatin remodeling at enhancers, promoters, and gene bodies and are associated with gene activation and repression (see for example10–15). In addition, members of the SWI/SNF complexes have been implicated in DNA repair, and genome instability16. Importantly, genes encoding subunits of the SWI/SNF complexes are mutated in about 20% of all human tumor samples, making them among the most frequently mutated complexes in cancer7,17–20. The mechanisms by which loss-of-function mutations in SWI/SNF complex subunits trigger tumor formation or affect tumor cell behavior is still a highly debated issue. Several data point to the pathological effects of aberrant residual SWI/SNF complexes as the cause of the potential selective advantage of SWI/SNF mutant cancer cells21–24.
The human SMARCA4 gene is frequently mutated in ovarian small cell carcinoma of the hypercalcemic type (in approx. 90% of the cases)25–27, and at much lower frequency in other cancer types28–32. In addition, SMARCA4 has been found to be silenced or mutated in a number of cancer cell lines33–35. Brg1 homozygous knockout mice die early during development; however, heterozygote mice or conditional inactivation of Brg1 in some adult tissues display increased tumor formation36–38. While SMARCA2 is not frequently mutated in tumors, it has been found to be silenced in a number of cancer cell lines39 and primary tumors33,40. Brm knockout mice develop normally, but Brm−/− embryonic fibroblasts present increased proliferation in vitro41. Furthermore, heterozygote and homozygote Brm mutants treated with carcinogens display increased tumor development39. Re-expression of SMARCA4 or SMARCA2 into cancer cell lines deficient for these proteins decreases cell proliferation34,42,43. Taken together, these results indicate that SMARCA4 and SMARCA2 have tumor suppressor activity. However, other recent reports point to essential roles of SMARCA4 and/or SMARCA2 in cell survival and proliferation in some types of cancers12,44–46, complicating our understanding of the role of these ATPases in cancer. In order to clarify this complex scenario, we have now used data from The Cancer Genome Atlas (TCGA) and other databases to investigate the levels of SMARCA4 and SMARCA2 mRNAs in several types of cancer. Notably, while SMARCA4 was mostly overexpressed in tumors, SMARCA2 expression decreased in tumors, as compared to normal tissue. A meta-analysis of prognosis data indicated that tumors with high SMARCA4 expression are mostly associated with poor prognosis, while tumors with high SMARCA2 expression are mostly associated with good prognosis. Analyzing liver hepatocellular carcinoma and kidney renal clear cell carcinoma TCGA cohorts, we found that high levels of SMARCA4 and SMARCA2 transcripts were inversely associated with survival prognosis, clinicopathological factors, and gene expression patterns, pointing to an inverse role of SMARCA4 and SMARCA2 in cancer.
Results
SMARCA4 is overexpressed, and SMARCA2 is underexpressed, in multiple types of tumors
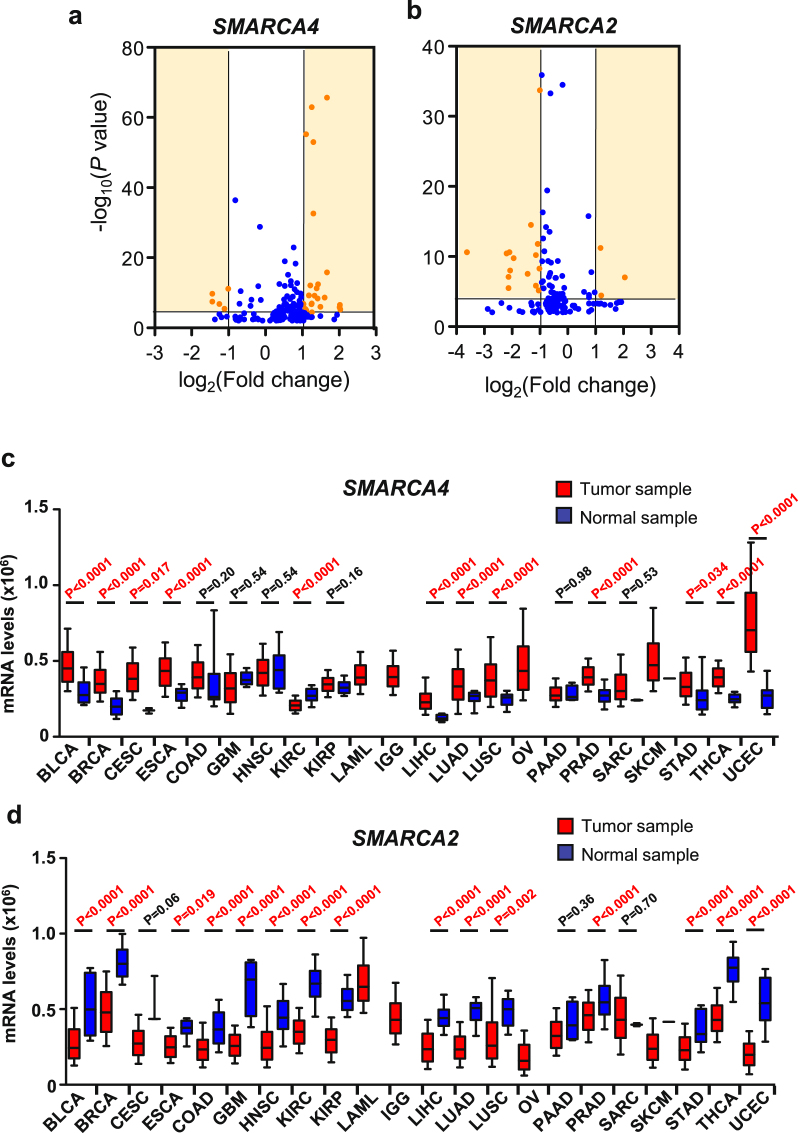
First we compared the levels of SMARCA4 and SMARCA2 transcripts in normal tissue with respect to tumor tissue in different types of tumors. For this, we performed a meta-analysis of microarray expression data from different studies collected form the ONCOMINE database47. In 130 out of the 161 datasets selected (see methods), SMARCA4 was found to be more highly expressed in the tumor samples than in the normal samples (Fig. 1a). Of the 32 datasets with highly significant changes (P ≤ 0.0001 and |lineal fold change (FC)| ≥ 2) (Fig. 1a; Supplementary Table S1), 26 (81%) presented higher levels of SMARCA4 transcript in tumor samples than in non-tumor samples. A similar analysis for SMARCA2 showed that its expression was reduced in 104 out of the 132 tumor datasets selected. Of the 16 datasets with highly significant changes (P ≤ 0.0001 and |FC| ≥ 2) (Fig. 1b and Supplementary Table S2), 13 (81%) presented lower levels of SMARCA2 transcript in tumor samples than in non-tumor samples. These data suggest that SMARCA4 gene is mostly overexpressed in tumors while, in contrast, SMARCA2 is mostly underexpressed in tumors. In order to corroborate these results, we collected total RNA-seq normalized data of 22 different types of tumors from The Cancer Genome Atlas (TCGA) consortium (Supplementary Table S3). Levels of SMARCA4 transcript were significantly higher in tumors than in normal tissue for 11 tumor types, and either unchanged or not quantifiable (due to lack of normal samples) in a further 10 tumor types (Fig. 1c). Only one type of tumor—kidney renal clear cell carcinomas (KIRC) — showed a higher expression of SMARCA4 in normal samples than in tumor samples. In stark contrast, SMARCA2 transcript levels were decreased in tumors compared to normal samples in 15 of the 22 tumor types analyzed and was not overexpressed in any type of tumor (Fig. 1d). These data confirm that SMARCA4 is mostly overexpressed in tumors, while SMARCA2 is mostly underexpressed in tumors.
Figure 1.
SMARCA4 is overexpressed, and SMARCA2 is underexpressed, in multiple types of tumors. (a,b) Volcano plots of SMARCA4 (a) or SMARCA2 (b) expression change (log2(FC)) in tumors with respect to normal samples versus significance (−log10(P value)), from different datasets. Data were obtained from ONCOMINE. Datasets with changes of P > 0.01 were not included. Highly significant changes (P ≤ 0.0001 and |FC| ≥ 2) are highlighted in orange and listed in Supplementary Tables S1 and S2. (c,d) Boxplot of levels of SMARCA4 (c) or SMARCA2 (d) mRNA (RNA-seq data) in normal or tumor samples of 22 different tumor cohorts from TCGA. Names and number of tumors and normal samples are listed in Supplementary Table S3. Significant Student’s t-test P value (P ≤ 0.05) are depicted in red.
We next investigated whether SMARCA4 overexpression occurs predominantly in tumors harboring SMARCA4 mutations as a possible consequence of a putative negative autoregulation. Using data on SMARCA4 mutations in 18 types of tumors obtained from TCGA through cBioPortal48, we found SMARCA4 to be mutated in either none or up to 8.5% of the samples, depending on the tumor type. SMARCA4 mutated tumors displayed similar level of accumulation of SMARCA4 mRNA as tumors harboring non-mutated SMARCA4 (Supplementary Fig. S1a). Similar results were found for SMARCA2 (Supplementary Fig. S1b). Taken together, these data demonstrate that SMARCA4 expression is upregulated and SMARCA2 is downregulated in most types of tumors irrespectively of the presence of mutations in the gene.
Inverse association for prognosis between high expression of SMARCA4 and SMARCA2
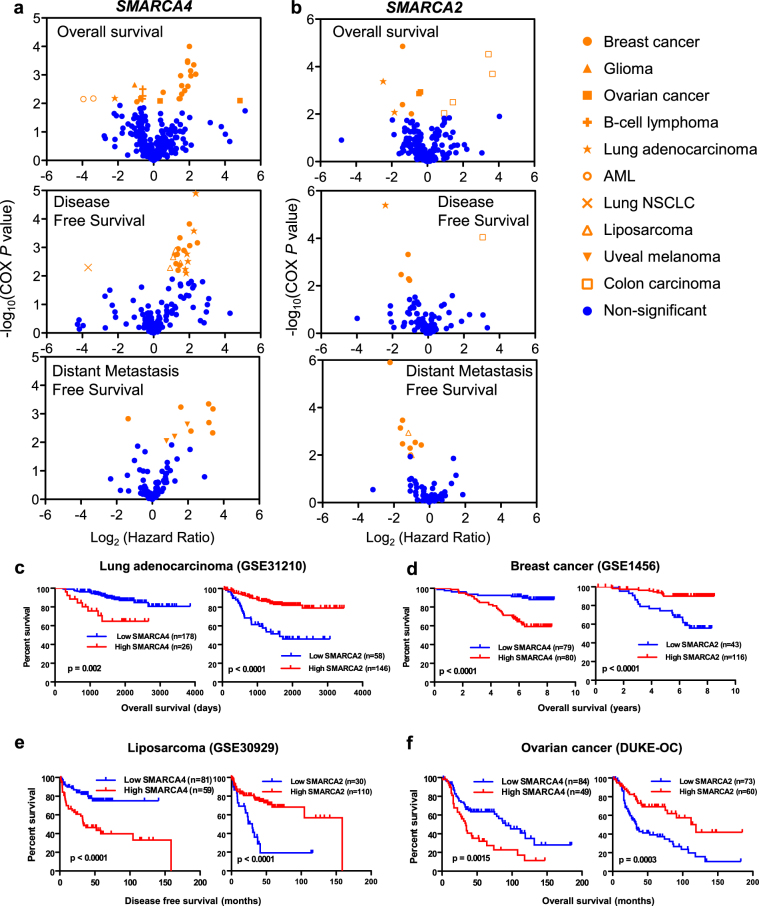
We next investigated whether having tumors with increased expression of SMARCA4 or SMARCA2 was linked to patient prognosis. For this, we performed a meta-analysis of data collected from the PrognoScan database49. This database allows systematic analysis of the prognostic value of the expression of a gene across a large collection of publicly available cancer microarray datasets. Correlation between gene expression and patient prognosis was evaluated using COX univariate analysis. Volcano plots of log2 hazard ratios (HR) versus −log10(COX P value) were drawn for every endpoint available (overall survival, disease-free survival, and distant metastasis-free survival). High expression of SMARCA4 was significantly associated (COX P ≤ 0.01) with a poor prognosis in breast and ovarian cancer, lung adenocarcinoma, liposarcoma and uveal melanoma datasets (Fig. 2a). In contrast, high expression of SMARCA2 was associated to good prognosis in breast and ovarian cancer, lung adenocarcinoma, and liposarcoma datasets (Fig. 2b). In fact, high expression of SMARCA2 was associated with poor prognosis only in colon carcinoma. Kaplan-Meier survival plots of patients from the same dataset, with high versus low expression of SMARCA4 and SMARCA2 are shown in Fig. 2c–f. These data indicate that, at least in some types of tumors, upregulation of SMARCA4 or SMARCA2 has opposite consequences in prognosis.
Figure 2.
Meta-analysis of prognosis consequences of SMARCA4 or SMARCA2 upregulation. (a,b) Cox regression analysis of the correlation between SMARCA4 (a) or SMARCA2 (b) tumor expression levels and patient survival in different studies (data collected from PrognoScan). Volcano plots of log2(HR) versus significance (−log10(COX P value)) from different datasets are shown. A positive log2(HR) value indicates that the analyzed risk increases when the level of gene expression increases, and thus the prognosis is worse. Conversely, a negative Log2(HR) value implies a better prognosis for patients with tumors with higher values of gene expression. Different survival endpoints (overall survival, disease-free survival, and distant metastasis–free survival) are shown in different graphics. Significant changes (COX P ≤ 0.01) are highlighted in orange. (c–f) Kaplan-Meier plots showing inverse prognosis behavior of SMARCA4 and SMARCA2 expression in the same studies. Data corresponding to four different types of tumors are shown: lung adenocarcinoma104, breast cancer105, liposarcoma106, and ovarian cancer107. GEO references of the data are provided when available. Long-rank test P values are provided. Patient tumors were divided into two expression groups (high and low) according to PrognoScan (see Methods).
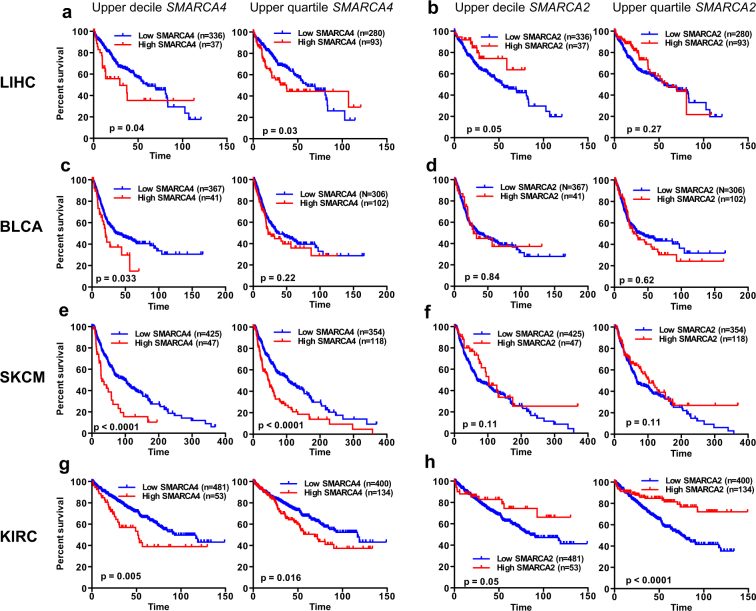
Next, we extended these studies to other types of tumors using TCGA data. Clinical data of cohorts of four different types of tumors were collected: liver hepatocellular carcinoma (LIHC)50, bladder urothelial carcinoma (BLCA)51, skin cutaneous melanoma (SKCM)52, kidney renal clear cell carcinoma (KIRC)53. Tumor collections were ranked according to the SMARCA4 or SMARCA2 mRNA levels (RNA-seq data). Then, survival of patients with expression values in the upper decile (first and third columns, Fig. 3) or upper quartile (second and fourth columns, Fig. 3) were compared with survival of the remaining patients. Analysis of these plots indicates that high expression of SMARCA4 is associated with poor prognosis in LIHC, BLCA, SKCM and KIRC (Fig. 3a,c,e,g). In clear contrast, high expression of SMARCA2 is associated with good prognosis in LIHC and KIRC (Fig. 3b,h) while in SKCM, prognosis improved but not significantly (Fig. 3f). Taken together, these data suggest that in most of the cohorts analyzed, high expression of SMARCA4 is associated with poor prognosis, while high expression of SMARCA2 is associated with good prognosis.
Figure 3.
High levels of SMARCA4 or SMARCA2 expression are frequently associated with the opposite prognosis. Kaplan Meier survival plots of patients with tumors that have high or low expression levels of SMARCA4 (a,c,e,g) or SMARCA2 (b,d,f,h) in liver hepatocellular carcinoma (LIHC), bladder urothelial carcinoma (BLCA), skin cutaneous melanoma (SKCM), or kidney renal clear cell carcinoma (KIRC) cohorts from TCGA. Patients with tumors that had expression values in the upper decile (left panels) or upper quartile (right panels) values were compared with the rest of the patients.
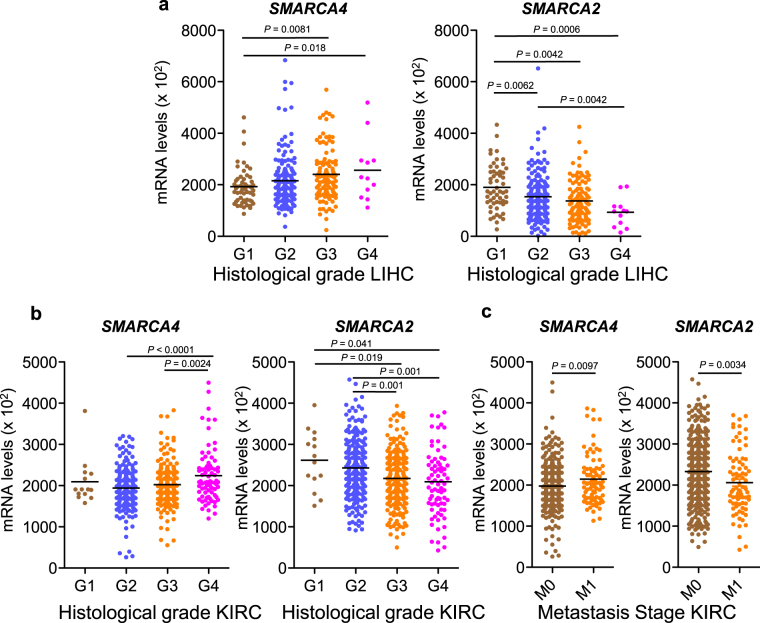
We further investigated the apparently opposite roles of SMARCA4 and SMARCA2 in the LIHC and KIRC tumors types. First we investigated whether expression levels of the SMARCA4 and SMARCA2 genes were associated to specific clinicopathological factors such as gender, age, tumor stage (T1 to T4 and stage I to IV), lymph node metastasis (N), distant metastasis (M), and differentiation (histological grade G1 to G4), obtained from TCGA. Consistently with the prognosis results, LIHC tumors with high levels of SMARCA4 expression (upper decile) presented a significant increased proportion of advanced stages, and poorly differentiated histology with respect to the rest of the LIHC tumors analyzed (Table 1). In contrast, tumors with high levels of SMARCA2 transcript (upper decile) presented increased proportion of well-differentiated tumors (Table 1). Further, increased undifferentiated histological grade is associated with a progressive increase of SMARCA4 and a decrease of SMARCA2 expression (Fig. 4a). Similarly, in KIRC tumors, high expression of SMARCA4 is associated with increased undifferentiated histological grade (Fig. 4b and Table 2), while high levels of SMARCA2 were associated with low tumor stages and well differentiated histology (Fig. 4b and Table 2). In addition, in KIRC tumors, high expression of SMARCA4 was strongly associated with the presence of metastasis (high proportion of N1, P = 0.035, and M1, P = 0.0009) (Table 2). In stark contrast, this trend was not observed in tumors with a high expression of SMARCA2. In fact, metastatic KIRC tumors (M1) presented significantly increased levels of SMARCA4 mRNA and decreased levels of SMARCA2 mRNA, with respect to non-metastatic tumors (Fig. 4c).
Table 1.
Association between SMARCA4 and SMARCA2 mRNA expression and clinicopathological factors in the liver hepatocellular carcinoma dataset from TCGA.
| Factor | SMARCA4 expression | SMARCA2 expression | ||||
|---|---|---|---|---|---|---|
| High expressiona n = 37 |
Lower expressionb n = 335 |
P value | High expressiona n = 37 |
Lower expressionb n = 335 |
P value | |
| Age | 57.4 ± 13.5 | 59.7 ± 13.5 | 0.30c | 60.8 ± 12.2 | 59.3 ± 13.6 | 0.49c |
| Gender | 0.01 d | 0.70d | ||||
| Male | 18 (48.6%) | 233 (69.6%) | 24 (64.9%) | 227 (67.8%) | ||
| Female | 19 (51.4%) | 102 (30.4%) | 13 (35.1%) | 108 (32.2%) | ||
| Tumor Stage (T) | 0.008 d | 0.29d | ||||
| T1 + T2 | 21 (56.8%) | 255 (76.8%) | 25 (67.6%) | 251 (75.6%) | ||
| T3 + T4 | 16 (43.2%) | 77 (23.2%) | 12 (32.4%) | 81 (24.4%) | ||
| Tumor Stage (S) | 0.010 d | 0.11d | ||||
| SI + SII | 19 (55.9%) | 239 (79.1%) | 22 (62.9%) | 236 (75.4%) | ||
| SIII + SIV | 15 (44.1%) | 75 (23.9%) | 13 (37.1%) | 77 (24.6%) | ||
| Lymph node metastasis | 0.014 d | 0.99d | ||||
| N0 | 27 (93.1%) | 226 (99.1%) | 25 (96.2%) | 228 (98.7%) | ||
| N1 | 2 (6.9%) | 2 (0.9%) | 1 (3.8%) | 3 (1.3%) | ||
| Metastasis Stage Code | 0.51d | 0.27d | ||||
| M0 | 30 (100%) | 235 (98.3%) | 24 (96.0%) | 243 (98.8%) | ||
| M1 | 0 (0.0%) | 4 (1.7%) | 1 (4.0%) | 3 (1.2%) | ||
| Histological grade | 0.004 d | 0.019 d | ||||
| G1 + G2 | 15 (41.7%) | 218 (65.9%) | 30 (81.1%) | 203 (61.5%) | ||
| G3 + G4 | 21 (58.3%) | 113 (34.1%) | 7 (18.9%) | 127 (38.5%) | ||
aDecile of the tumor population with higher levels of SMARCA4 or SMARCA2 mRNA.
bRest of the tumors not included in the high expression decile.
cStudent’s t-test.
dChi-square test. Significant P values (P ≤ 0.05) are depicted in bold.
Figure 4.
Correlation between SMARCA4 or SMARCA2 expression levels and clinicopathological factors. (a) Boxplot of levels of SMARCA4 (left panel) and SMARCA2 (right panel) transcript (normalized RNA-seq data) in LIHC tumors with different histological grades (G1 to G4). (b) Boxplot of transcript levels of SMARCA4 (left panel) or SMARCA2 (right panel) in KIRC tumors with different histological grades (G1 to G4). (c) Boxplot of transcript levels of SMARCA4 (left panel) or SMARCA2 (right panel) in KIRC tumors with different metastasis stages (M0 or M1). See Methods for description of G and M grading.
Table 2.
Association between SMARCA4 and SMARCA2 mRNA expression and clinicopathological factors in the renal clear cell carcinoma dataset from TCGA.
| Factor | SMARCA4 expression | SMARCA2 expression | ||||
|---|---|---|---|---|---|---|
| High expressiona n = 54 |
Lower expressionb n = 479 |
P value | High expressiona n = 54 |
Lower expressionb n = 479 |
P value | |
| Age | 58.48 ± 11.0 | 60.84 ± 12.2 | 0.15c | 57.6 | 60.9 | 0.06c |
| Gender | 0.037 d | 0.99d | ||||
| Male | 28 (51.9%) | 317 (66.2%) | 35 (64.8%) | 310 (64.7%) | ||
| Female | 26 (48.1%) | 162 (33.8%) | 19 (35.2%) | 169 (35.3%) | ||
| Tumor stage | 0.16d | 0.028 d | ||||
| T1 + T2 | 30 (55.6%) | 312 (65.1%) | 42 (75.9%) | 300 (62.6%) | ||
| T3 + T4 | 24 (44.4%) | 167 (34.9%) | 12 (24.1%) | 179 (37.4%) | ||
| Tumor stage (S) | 0.076d | 0.019 d | ||||
| SI + SII | 27 (50.0%) | 297 (62.4%) | 41 (75.9%) | 283 (59.5%) | ||
| SIII + SIV | 27 (50.0%) | 179 (37.6%) | 13 (24.1%) | 193 (40.5%) | ||
| Lymph node metastasis | 0.035 d | 0.06d | ||||
| N0 | 30 (85.7%) | 209 (95.0%) | 26 (92.9%) | 213 (93.8%) | ||
| N1 | 5 (14.3%) | 11 (5.0%) | 2 (7.1%) | 14 (6.2%) | ||
| Metastasis Stage Code | 0.0009 d | 0.24d | ||||
| M0 | 34 (68.0%) | 387 (86.0%) | 45 (86.5%) | 376 (83.9%) | ||
| M1 | 16 (32.0%) | 63 (14.0%) | 7 (13.5%) | 72 (16.1%) | ||
| Histologic grade | 0.043 d | 0.008 d | ||||
| G1 + G2 | 17 (32.7%) | 224 (47.5%) | 33 (63.5%) | 209 (44.2%) | ||
| G3 + G4 | 35 (66.0%) | 248 (52.5%) | 19 (36.5%) | 264 (55.8%) | ||
aDecile of the tumor population with higher levels of SMARCA4 or SMARCA2 mRNA.
bRest of the tumors not included in the high expression decile.
cStudent’s t-test.
dChi-square test. Significant P values (P ≤ 0.05) are depicted in bold.
Transcriptome changes associated with SMARCA4 or SMARCA2 upregulation in liver hepatocellular carcinoma
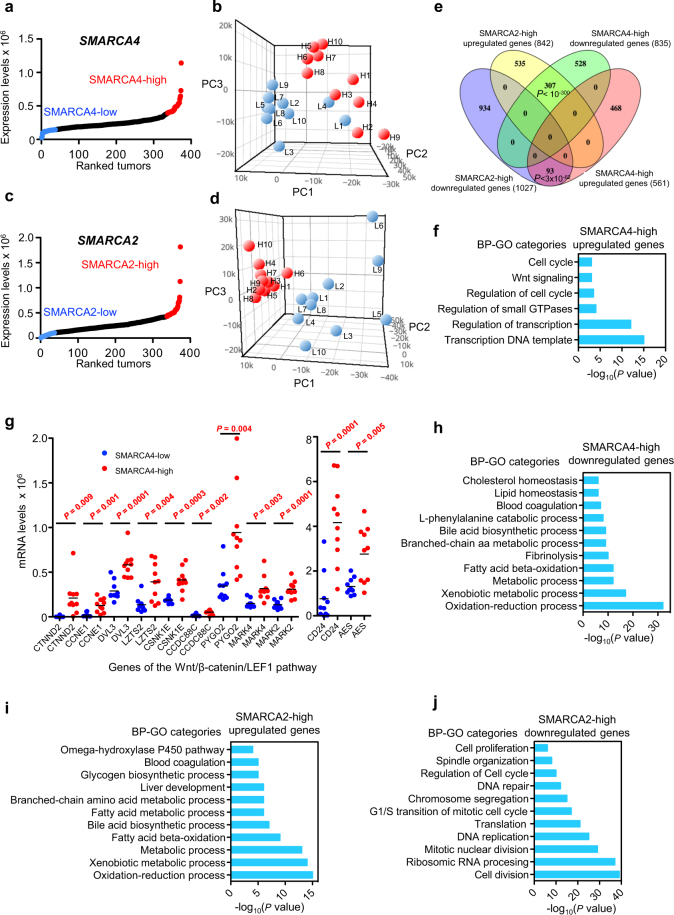
We next investigated the gene expression patterns that characterize LIHC tumors with high expression of SMARCA4 or SMARCA2. For that, LIHC tumors were ranked according to the level of SMARCA4 mRNA, and ten tumors were randomly selected from the upper decile (SMARCA4-high) or from the lower decile fractions (SMARCA4-low) (Fig. 5a; Supplementary Table S4). RNA-seq transcriptomic data of 60,483 genes from the 20 selected tumors were collected from TCGA and subjected to unsupervised principal component (PC) analysis. PC analysis differentiated almost all SMARCA4-low from SMARCA4-high tumors (only two tumors, L1 and L4, had an intermediate pattern), suggesting that the level of SMARCA4 expression characterizes different subtypes of LIHC tumors (Fig. 5b). We then selected genes that were differentially expressed (P ≤ 0.01 and |FC| ≥ 2) in the SMARCA4-high versus the SMARCA4-low collection of tumors. Of the 1396 differentially expressed genes, 561 were upregulated and 835 were downregulated in SMARCA4-high tumors (Supplementary Fig. S2; Supplementary Table S5). A similar analysis was performed for SMARCA2-high versus the SMARCA2-low tumors (Fig. 5c; Supplementary Table S4). In this case, PC analysis clearly differentiated all SMARCA2-high from the SMARCA2-low tumors, being all SMARCA2-high tumors very closely related according to the three PCs analyzed (Fig. 5d). Differential expression analysis showed that 842 genes were significantly (P ≤ 0.01 and |FC| ≥ 2) upregulated and 1027 genes were downregulated, in SMARCA2-high versus SMARCA2-low tumors (Supplementary Fig. S2; Supplementary Table S6). Interestingly, the genes that were downregulated or upregulated in SMARCA4-high tumors strongly overlapped with the genes that were upregulated or downregulated, respectively, in SMARCA2-high tumors (Fig. 5e), demonstrating that these types of tumors not only have inverse prognosis but also opposite gene expression patterns.
Figure 5.
Transcriptome changes associated with SMARCA4 or SMARCA2 upregulation in LIHC. (a,c) Ranking (x-axis) of LIHC tumor samples according to SMARCA4 (a) or SMARCA2 (c) expression (normalized RNA-seq data) (y-axis). Dots corresponding to upper and lower deciles are depicted in red and blue, respectively. (b) Principal component (PC) analysis of transcriptomic data from ten SMARCA4-high (red) or ten SMARCA4-low (blue) tumors. (d) PC analysis of transcriptomic data from ten SMARCA2-high (red) or ten SMARCA2-low (blue) tumors. (e) Venn diagram showing overlap between SMARCA2-high upregulated and SMARCA4-high downregulated genes, and between SMARCA2-high downregulated and SMARCA4-high upregulated genes. Significance of overlap based on the hypergeometric test is provided. (f) Biological process (BP)-GO categories enriched in SMARCA4-high upregulated genes. (g) Expression levels (normalized RNA-seq data) of genes in the Wnt/β-catenin/LEF1 signal transduction pathway in SMARCA4-high and SMARCA4-low LIHC tumors. Student’s t-test P values are shown. (h) BP-GO categories enriched in SMARCA4-high downregulated genes. (i,j) BP-GO categories enriched in SMARCA2-high upregulated (i) and SMARCA2-high downregulated (j) genes. (f,h–j) Significance of the enrichment are presented as the P values of the hypergeometric test (−log10 transformed).
Genes upregulated in SMARCA4-high tumors were enriched in regulation of transcription, cell cycle, DNA replication, and Wnt signaling pathways and functional categories (Fig. 5f; Supplementary Figs S3a and S4a). Furthermore, DNA binding sites for the LEF1 (P = 8.9 × 10−18), MAZ (P = 1.7 × 10−12), SP1 (P = 1.8 × 10−10) and E2F (P = 2.7 × 10−10) transcription factors were strongly overrepresented in the promoter regions of upregulated genes. In agreement with the enrichment in Wnt signaling categories and LEF1 DNA binding sites, several genes related to this pathway were activated (Fig. 5g). Genes downregulated in SMARCA4-high tumors were enriched in lipid and amino acids metabolism, xenobiotic metabolism, blood coagulation and aerobic respiration categories and pathways (Fig. 5h; Supplementary Figs S3b and S4a). All of these processes are important liver functions carried out by differentiated hepatocytes54. Consistently, promoters of downregulated genes were enriched in binding sites for typical liver transcription factors such as HNF1 (P = 9.1 × 10−11), FOXO4 (P = 5.9×10−8), and HNF4 (P = 6.4×10−7). These data suggest that SMARCA4-high tumors present a strong decrease of hepatocytes-specific functions, which is in agreement with the high proportion of undifferentiated cells (G3 + G4 histological grade) and the poor prognosis observed in these tumors (Table 1; Fig. 3a).
In contrast to SMARCA4-high tumors, SMARCA2-high tumors had upregulated gene sets prominently involved in hepatocytes-specific functions such as fatty acid metabolism, amino acid metabolism, drugs and xenobiotic metabolism, blood coagulation, and respiration categories and pathways (Fig. 5i; Supplementary Figs S3c and S4b). For instance, genes encoding typical hepatic enzymes such as tyrosine aminotransferase (TAT) and alcohol dehydrogenases 1B and 1 C (ADH1B, ADH1C) were increased 43.2-, 42.3- and 14.9-fold, respectively. In addition, genes downregulated in SMARCA2-high tumors were strongly enriched in ribosome RNA processing, translation, cell cycle, DNA-replication, and mitosis-related functions and pathways (Fig. 5j; Supplementary Figs S3d and S4b), and often presented E2F (P = 1.05×10−26), MYC (P = 3.9×10−16) and ELK1 (P = 1.2 × 10−11) binding sites, suggesting a reduced proliferation of these tumor cells. In sum, these data suggest that SMARCA2-high tumors maintain a high differentiation stage with low levels of proliferation, which is consistent with the good prognosis for patients with tumors with high levels of SMARCA2 expression.
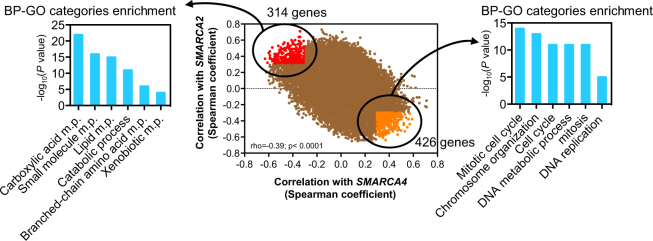
In order to further characterize the roles of SMARCA4 and SMARCA2 in liver hepatocellular carcinoma, we have performed a genome-wide co-expression analysis using all available LIHC tumors from the TCGA cohort. We calculate Spearman correlation coefficients (rho) between the expression levels of SMARCA4 or SMARCA2 and 22,300 genes (data collected from cBioportal). A scatter plot was drawn with each dot corresponding to a gene, and the x and y coordinates as the Spearman correlation coefficient with SMARCA4 and with SMARCA2, respectively. This showed a negative correlation (rho = −0.39; P < 0.0001) between Spearman coefficients, indicating that most of the genes whose expression is positively correlated with SMARCA4 are negatively correlated with SMARCA2, and vice-versa (Fig. 6). We next analyzed the gene ontology (GO) of the genes that are most robustly co-regulated in an opposite way with SMARCA4 and SMARCA2 (rho ≥0.3 and ≤−0.3). Genes positively coexpressed with SMARCA4 and negatively coexpressed with SMARCA2 (Supplementary Table S7) were enriched in cell cycle (CCNB1, CCNE1, CDK1, E2F1), mitosis (PLK, AURKB, CDC20, CDC25A) and DNA replication (POLD1, RFC4) GO categories. In contrast, genes positively coexpressed with SMARCA2 and negatively coexpressed with SMARCA4 (Supplementary Table S7) were enriched in liver metabolism functions, such as lipid metabolism (ACADL, ACSL1, LIPG), amino acid metabolism (TAT, BCKDHB, PAH, IDH1), xenobiotic detoxification (CYP3A4, CYP4V2, CYP8B1), and blood coagulation (F8, F11) categories. This analysis confirms that high expression of SMARCA4 or SMARCA2 characterizes types of LIHC tumors with opposite patterns of gene expression.
Figure 6.
Analysis of genes co-expressed with SMARCA4 and SMARCA2 in LIHC. Scatter plot with each dot corresponding to a gene, with the x and y coordinates as the Spearman coexpression correlation coefficient of the gene with SMARCA4 and SMARCA2, respectively (central panel). Spearman correlation coefficient (rho) and P values are shown. Genes that were more positively correlated with SMARCA4 (rho ≥ 0.3) and negatively correlated with SMARCA2 (rho ≤ −0.3) are depicted in orange. Genes that were more negatively correlated with SMARCA4 (rho ≤ −0.3) and positively correlated with SMARCA2 (rho ≥ 0.3) are depicted in red. Enrichment of GO categories is given for genes negatively correlated with SMARCA4 and positively correlated with SMARCA2 (left panels) and for genes positively correlated with SMARCA4 and negatively correlated with SMARCA2 (right panels). m.p. metabolic process.
Transcriptome changes associated with SMARCA4 or SMARCA2 upregulation in kidney renal clear cell carcinoma
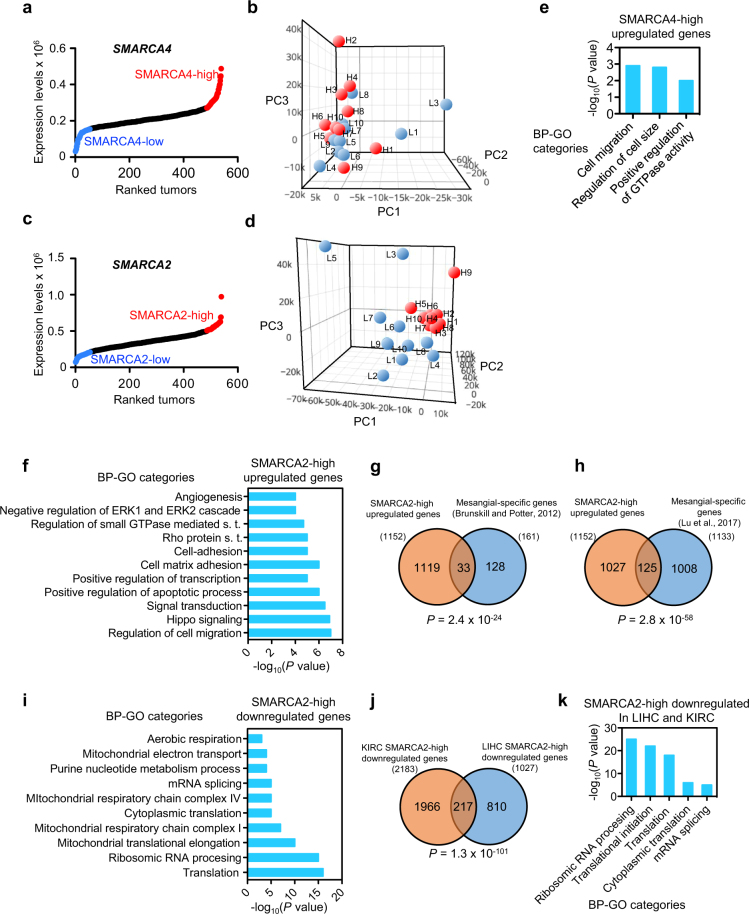
A similar gene expression analysis as for LIHC tumors (above) was then performed for KIRC tumors. For this, ten tumors of each type: SMARCA4-high, SMARCA4-low, SMARCA2-high and SMARCA2-low, were selected from the KIRC cohort (Fig. 7a,c; Supplementary Table S4), and RNA-seq transcriptomic data from 60,483 genes were collected from TCGA. PC analysis of whole transcriptomic data was unable to discriminate between SMARCA4-high and SMARCA4-low tumors (Fig. 7b). However, PC analysis clearly differentiated SMARCA2-high from SMARCA2-low tumors (Fig. 7d), as SMARCA2-high tumors were very closely grouped with respect to the three PCs analyzed.
Figure 7.
Transcriptome changes associated with SMARCA4 or SMARCA2 upregulation in KIRC. Ranking (x-axis) of KIRC tumor samples according to SMARCA4 (a) or SMARCA2 (c) expression (normalized RNA-seq data) (y-axis). Dots corresponding to the upper or lower decile are depicted in red or blue, respectively. (b) PC analysis of transcriptomic data from ten SMARCA4-high (red) and ten SMARCA4-low (blue) tumors. (d) PC analysis of transcriptomic data from ten SMARCA2-high (red) and ten SMARCA2-low (blue) tumors. (e,f) BP-GO categories enriched in SMARCA4-high (e) or SMARCA2-high (f) upregulated genes. s.t. signal transduction. (g,h) Overlapping between SMARCA2-high upregulated genes and mesangial cells-specific genes from two independent studies66: (g) and67 (GSE92650) (h). (i) BP-GO categories enriched in SMARCA2-high downregulated genes. (j) Overlap between SMARCA2-high downregulated genes from LIHC and KIRC tumors. (g,h,j) P values of the hypergeometric test. (k) BP-GO categories enriched in genes overlapping between SMARCA2-high upregulated genes from LIHC and from KIRC tumors. (e,f,i,k) Significance of the enrichments are given as the P values of the hypergeometric test (−log10 transformed).
Only 140 genes were differentially expressed (P ≤ 0.01 and |FC| ≥ 2) in the SMARCA4-high versus the SMARCA4-low collection of tumors, of which 69 were upregulated and 71, downregulated (Supplementary Fig. S2; Supplementary Table S8). Upregulated genes were significantly enrichment in categories related to regulation of small GTP hydrolases (GTPases) (Fig. 7e; Supplementary Fig. S5a). Three genes encoding guanine nucleotide exchange factors (GEF) were upregulated: ARHGEF18, RASGEF1A, and VAV2. While ARHGEF18 and VAV2 are Rho GEFs, RASGEF1A is a regulator of RAP2. GEFs promote the transition of small GTPases from the inactive (GDP bound) to the active (GTP bound) state during signal transduction. Rho and RAP GTPases play essential roles in the regulation of cell morphology, cytokinesis, cell adhesion, and cell migration, and their activation and overexpression have been associated to metastasis55,56. Further, high expression of VAV257, ARHGEF1858 and RASGEF1A59 has also been linked to metastasis, which is consistent with the high proportion of metastasis (M1 code) and metastatic lymph nodes (N1 code) we observed in SMARCA4-high tumors (Table 2). SMARCA4-high tumors also display high expression of PAX8, a well-known marker for primary and metastatic renal clear cell carcinomas60,61.
We observed 1152 genes to be upregulated, and 2183 genes downregulated, in the SMARCA2-high with respect to the SMARCA2-low tumors (Supplementary Fig. S2; Supplementary Table S9). SMARCA2-high upregulated genes were enriched in regulation of transcription, regulation of cell migration, regulation of small GTPases, focal adhesion, negative regulation of ERK1/2 cascade, and tight junction categories and pathways (Fig. 7f; Supplementary Figs S5b and S6a). Interestingly, several genes encoding Rho GTPase activating proteins (GAPs) were found to be upregulated in SMARCA2-high tumors, such as DLC1 (FC = 3.8) NF1 (FC = 2.3), ARHGAP19 (FC = 2.1), ARHGAP31 (FC = 3.0). This is in clear contrast with the presence of high levels of Rho GEFs in SMARCA4-high tumors. DLC162,63 and NF164 are well-known tumor suppressors, which is consistent with the good prognosis of the SMARCA2-high tumors. Increased expression levels of genes encoding cell-cell contact molecules (tight, GAP and adherens junctions) were also observed, including the TJP1, JMY, JAM2, JAM3, GJA1, and OCLN genes, suggesting a marked epithelial or endothelial phenotype of the SMARCA2-high tumors. Several genes encoding markers of endothelial cells were upregulated, such as PCAM1, VWF, CD34, NRP1, TEK, and FLT1, consistent with the fact that the renal glomerulus is mostly formed by three types of cells: endothelial cells, podocytes and mesangial cells65. Interestingly, we also observed a significant overlap between mesangial-expressed genes66,67 and genes upregulated in SMARCA4-high tumors (Fig. 7g,h). The fact that SMARCA2-high tumors express high levels of markers of glomerulus cell types is in agreement with the high differentiation (e.g., low histologic grade) of these tumors (Table 2).
Genes downregulated in SMARCA2-high tumors were strongly enriched in categories related to both cytosolic and mitochondrial ribosomal proteins, translation, and mitochondrial respiration electron transport (Fig. 7i; Supplementary Figs S5b and S6b). Promoters of downregulated genes were very significantly enriched in binding sites for ELK1 (P = 6.6 × 10−45), NRF1 (P = 1.4 × 10−21), NRF2 (P = 1.5 × 10−9) and MYC (P = 6.3 × 10−15). NRF1 and NRF2 have important functions controlling cell growth, respiration, mitochondrial DNA transcription and replication68. The downregulation of mitochondrial respiration suggested that SMARCA2-high tumors might present a strong Warburg effect69. However, none of the ten glycolysis genes (gene set M15109 from MSigDB) were upregulated in SMARCA2-high tumors, in fact the expression levels of GAPDH (FC = 0.47; P = 1.3×10−5) and PFKFB4 (FC = 0.41; P = 0.0007) decreased, suggesting that SMARCA2-high tumors have a reduced energetic metabolism. Notably, analyzing for similarity between the transcription patterns of LIHC SMARCA2-high and KIRC SMARCA2-high tumors revealed a very significant overlap between downregulated genes in these two types of tumors (Fig. 7j). Most overlapping genes encoded ribosomal proteins and proteins related to translation and ribosome biogenesis (Fig. 7k). These data suggest that LIHC and KIRC tumors with high expression of SMARCA2 have reduced translation and, therefore, probably a reduced cell growth.
Discussion
SMARCA4 is frequently upregulated in tumors
A role of SWI/SNF complexes as tumor suppressors is widely accepted, mostly based on the fact that genes encoding SWI/SNF subunits are mutated in a wide-ranging proportion of tumors18,20. Thus, SMARCA4 is frequently mutated (more than 90% of the cases) in ovarian small cell carcinoma of the hypercalcemic type25–27. However, several studies and inspection of the TCGA data indicate that, in most of the tumor types SMARCA4 mutations vary between 0% and 15% of the cases18,20,25–32. Tumor suppressor genes are normally either mutated or downregulated in tumor tissues70. However, we now show that SMARCA4 is frequently overexpressed in tumors. Furthermore, we show that SMARCA4 upregulation is associated with a poor prognosis in published datasets for breast and ovarian cancer, lung adenocarcinoma, liposarcoma, and uveal melanoma and in the SKCM, LIHC, BLCA, and KIRC TCGA cohorts, indicating that high expression of SMARCA4 can be used as a prognosis marker for these types of tumors. Consistently, loss of expression of SMARCA4 protein has been recently associated to improved prognosis in clear cell renal cell carcinoma71. Increased expression of SMARCA4 has been previously reported in several types of tumors45,46,72–77. In addition, several studies have shown that SMARCA4 is required for tumor cell proliferation12,44–46. Furthermore, we find that high levels of SMARCA4 expression are associated with an advanced tumor stage and histological grade in LIHC, and with increased metastasis in KIRC. Taken together, these data suggest that, at least for several types of cancers, high expression of SMARCA4 confers a selective advantage to tumor cells. This is, therefore, not consistent with a general role of SMARCA4 as a tumor suppressor. A context-dependent dual role of SMARCA4 in cancer has been also proposed by Dr. Imbalzano and collaborators78. A growing number of genes play both tumor suppressor or oncogenic roles in different tissue, tumor types or experimental settings79. Therefore, our data are not incompatible with a role of SMARCA4 as a tumor suppressor when it is mutated in certain types of tumors, probably due to the pathological activity of aberrant residual SWI/SNF complexes.
What is the mechanism by which increased levels of SMARCA4 are important for cancer development? The answer is still unclear but probably it is dependent on the cancer type. SMARCA4 has been shown to promote breast cancer by reprogramming lipid synthesis45 and to be required for maintaining repopulation of hematopoietic stem cells in leukemia44. In fact, a role of SMARCA4 in regulation of stem cells pluripotency has been well characterized11,80, and SMARCA4 is highly expressed in stem cells81. Therefore, it is possible that SMARCA4 plays an essential role in the maintenance of cancer stem cells. The role of the Wnt/β-catenin/LEF1 pathway in activation of hepatic cancer stem cells in hepatocellular carcinoma and during liver regeneration has been well characterized82–84. Interestingly, we observed that LIHC SMARCA4-high tumors presented increased levels of several genes of the Wnt/β-catenin/LEF1 pathway. This is also consistent with the relative undifferentiated state of these tumors, according to gene expression pattern and histological grade. In addition, we show that LIHC SMARCA4-high tumors had high expression of positive regulators of cell cycle progression and mitosis, such as cyclins, mitotic kinases, and DNA replication factors, which also suggests a positive correlation between levels of SMARCA4 and proliferation in LIHC. Consistently, Kaufmann et al., recently showed that knockdown of SMARCA4 impairs proliferation and decreases cyclin B and cyclin E expression in hepatocellular carcinoma cell lines77. A role of SWI/SNF complexes containing SMARCA4 in positive regulation of cell cycle genes85 has been previously described.
In KIRC SMARCA4-high tumors, we did not find increased expression of the Wnt/β-catenin/LEF1 pathway or cell cycle genes, illustrating the absence of similarity between SMARCA4-high tumors of different origins. However, we found that KIRC SMARCA4-high tumors presented a high proportion of metastasis. SMARCA4-high tumors displayed high expression of the RhoA GEFs ARHGEF18 and VAV2, which are involved in activation of RhoA small GTPase. The RhoA signaling pathway and ARHGEF18 and VAV2 proteins have been implicated in metastasis formation56–58. Interestingly, RhoA signaling activation was reported upon SMARCA4 re-expression in SMARCA4-deficient human adrenal adenocarcinoma SW13 cells86.
SMARCA2 is frequently downregulated in tumors
In contrast to SMARCA4, SMARCA2 expression was strongly downregulated in most cancer types, which is consistent with a role as tumor suppressor of this protein. SMARCA2 levels were correlated with good prognosis in published datasets for breast and ovarian cancer, lung adenocarcinoma, and liposarcoma, and in the LIHC and KIRC TCGA cohorts. In addition, high levels of SMARCA2 expression were associated with a low tumor stage and well-differentiated tumors in LIHC and KIRC. SMARCA2 is not frequently mutated in tumors but gene silencing in tumor cell lines has been reported33,39,43. Several experimental data support a role of SMARCA2 as a tumor suppressor. Brm−/− mouse embryonic fibroblasts present increased proliferation and have lost inhibition of growth by cell-cell contact41. Additionally, heterozygote and homozygote Brm mutant mice treated with carcinogens have increased tumor development39,87. Expression of SMARCA2 is negatively regulated by mitogenic stimulation and Ras and ERK signaling, and restoration of SMARCA2 levels leads to reversion of the transformed phenotype42,43,88. Finally, SMARCA2 is not expressed in stem cells or during early development until the stage of blastocyst, and its levels increase during stem cells differentiation and during late development41,81. Recent data also show that SMARCA2 is required for cell cycle arrest during myoblast differentiation89. Taken together, these data suggest that a reduced level of SMARCA2 expression confers a selective advantage for many types of tumor cells. In agreement with this, LIHC and KIRC SMARCA2-high tumors form a coherent and well-defined subtype of tumors, with high differentiation according to gene expression patterns and histological grade and with low expression of cell cycle genes (for LIHC) and low expression of ribosomal and translation genes (for both LIHC and KIRC). A reduction in the levels of SMARCA2 transcript in a cohort of hepatocellular carcinomas has been previously reported90. In this study, SMARCA2 protein expression was lost in nine of 40 tumors and patients with these tumors presented a poor overall survival. Similarly, decrease of overall survival in SMARCA2 negative tumors has been also recently reported in clear cell renal cell carcinoma but only when levels of PBRM1 protein, another subunit of the SWI/SNF complex were also reduced71,91. These data suggest that SMARCA2 expression is a good marker for characterizing LIHC and KIRC prognosis.
Do SMARCA4 and SMARCA2 play antagonistic roles in cancer?
Our data demonstrate that levels of SMARCA4 and SMARCA2 expression correlate with opposite prognosis in several types of tumors and, in addition, with opposing clinicopathological factors and gene expression patterns in LIHC and KIRC tumors. Whether SMARCA4 and SMARCA2 expression are the cause or the consequence of differences in tumors is not yet clear. However, the facts that SMARCA4 expression is mostly associated to cell types that constantly undergo proliferation or self-renewal81,92 while SMARCA2 is absent from stem cells and inversely correlated with proliferation in several types of cells42,88,89,92, suggest the attractive possibility that the SWI/SNF complexes use a different ATPase, or a different ratio of ATPases, for proliferating-undifferentiated versus quiescent-differentiated conditions. How this equilibrium would be controlled is unclear. However, it has been demonstrated that a decrease in the protein level of one of the ATPases of the SWI/SNF complexes causes an increase of the level of the paralogous ATPase as well as its replacement in the complexes41,93, suggesting that the changes of the mRNA levels in tumors that we describe in this work can alter the composition of the complexes. Finally, it has been proposed that the SMARCA4 and SMARCA2 ATPases are appropriate targets for anticancer drugs design94. The antagonistic behavior uncovered in our work should be taken into account to design specific drugs that specifically target one but not the other ATPase.
Methods
Data collection and analysis of SMARCA4 and SMARCA2 levels
For meta-analysis of SMARCA4 and SMARCA2 transcript levels in normal and tumor samples data were collected from the cancer microarray expression database ONCOMINE 4.547 (https://www.oncomine.org/). Those datasets in which changes of expression between normal and tumor tissue were significant, with a P value ≤ 0.01 (Student’s t-test), and which ranked in the top 10% of the more significant changes, were selected. Volcano plots of −Log10(P value) versus log2(FC) were then generated.
Expression of SMARCA4 and SMARCA2 was also analyzed in 22 cohorts of different types of tumors from The Cancer Genome Atlas (TCGA) consortium. SMARCA4 and SMARCA2 RNA-seq mRNA expression data (FPKM-UQ normalized) in tumors and the corresponding available normal tissue samples were collected from TCGA (https://cancergenome.nih.gov/) though the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov). Types of tumors as well as the number of tumors and normal samples are provided in Supplementary Table S3.
All methods and use of data were carried out in accordance with relevant guidelines and regulations of the corresponding databases. No experiments were performed using human samples.
Analysis of prognosis
Meta-analyses of the association of SMARCA4 and SMARCA2 expression levels with survival outcomes were performed using data collected from the PrognoScan database (http://www.abren.net/PrognoScan/)49. COX regression analysis95 data (HR and COX P value) were downloaded and used to construct volcano plots of log2(HR) versus −log10(COX P value) for every endpoint available (overall survival, disease-free survival and distant metastasis-free survival). Only studies with COX P values ≤ 0.01 were considered as significant. For Kaplan-Meier plots of Fig. 2c–f, patients were divided into two (high and low) groups according on the expression of SMARCA4 or SMARCA2 in the tumors. The optimal cut-point that gave the most pronounced corrected P value (in the log-rank test) between the two groups was provided by the PrognoScan database.
Association between SMARCA4 or SMARCA2 expression levels and prognosis was also analyzed in four cohorts of TCGA: Liver hepatocellular carcinoma (LIHC), bladder urothelial carcinoma (BLCA), skin cutaneous melanoma (SKCM), and kidney renal clear cell carcinoma (KIRC). Overall survival data of patients were collected from TCGA, and Kaplan-Meier plots were performed in Prism 5 (GraphPad). Significance was determined using log-rank test.
Clinical data
Clinicopathological data of patients in the LIHC and KIRC cohorts were obtained from TGCA. For tumor description, the Tumor-Node-Metastasis (TNM) staging system (www.cancerstaging.org/) was used, whereby T followed by a number (1–4) describes the size of the tumor (with T4 being the largest); N followed by 1 or 0 indicates whether lymph nodes have metastasis or not, respectively; and M followed by 1 or 0 indicates whether the tumor has metastasized or not, respectively. Histopathologic grade G followed by a number (1–4) was also considered: G1, well differentiated; G2, moderately differentiated; G3, poorly differentiated and G4, undifferentiated tumor. We also considered the roman numeral stage annotation (S) from I to IV, with each number corresponding approximately to a combination of the TNM numbers. No subdivisions of stages were used (e.g., T1a, T1b, and T1c were considered as T1). To determine significance of differences between groups either Student’s t-test or Chi-square test with confidence interval of 95% were computed, using Prism 5 (GraphPad).
Principal component and differential expression analysis
To characterize gene expression changes between tumors with high or low levels of SMARCA4 or SMARCA2, all analyzed LIHC or KIRC TCGA tumors were ranked according to their level of SMARCA4 mRNA or SMARCA2 mRNA, respectively. Ten tumors were then randomly selected from the upper decile (SMARCA4-high or SMARCA2-high) or from the lower decile (SMARCA4-low or SMARCA2-low) for each tumor type. These were used to generate following sets of ten tumors: SMARCA4-high LIHC, SMARCA4-low LIHC, SMARCA2-high LIHC, SMARCA2-low LIHC, SMARCA4-high KIRC, SMARCA4-low KIRC, SMARCA2-high KIRC, SMARCA2-low KIRC. The reference numbers of the TCGA tumors used is provided in Supplementary Table S4. The expression patterns of SMARCA4-high versus SMARCA4-low, and SMARCA2-high versus SMARCA2-low, in both types of tumors were then subjected to PC analysis. For this, RNA-seq expression data (FPKM-UQ normalized) of 60,483 Ensembl reference genes from the 20 compared samples were centered on the median (z-scores), and non-expressed genes in any of the samples were removed. Data were then subjected to unsupervised PC analysis using default parameters in MultiExperiment viewer (MeV) 4.8.1 software96. Data of the three PCs (PC1, PC2 and PC3) were then represented in 3D scatter plots using Plotly 2.0(https://plot.ly/create/).
Differential expression analyses between sets of tumors (SMARCA4-high LIHC versus SMARCA4-low LIHC; SMARCA2-high LIHC versus SMARCA2-low LIHC; SMARCA4-high KIRC versus SMARCA4-low KIRC; SMARCA2-high KIRC versus SMARCA2-low KIRC) were performed using RNA-seq expression data (FPKM-UQ normalized) of 60483 Ensemble genes and standard methods97. Unpaired two-samples Students t-test with unequal variances was used to compute P values. Differentially expressed genes were considered to be significant when P ≤ 0.01 and |FC| ≥ 2. Expression values of differentially expressed genes were then centered on the median (z-scores), and heat maps were produced using MeV 4.8.1 software96. Unsupervised hierarchical clustering analysis (HCA) of the differentially expressed genes was also performed in MeV 4.8.1. Clustering was done with complete linkage and Euclidean distance. Venn diagrams were performed in Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html). To test the significance of overlap in Venn diagrams, the hypergeometric tests were performed in R, using the dhyper function from the stats package. Population size was considered to be 60,483, the total number of genes for which RNA-seq data were available in TCGA.
Functional enrichment analysis
Gene ontology (GO) functional categories were analyzed using DAVID98 or WebGestalt99 software packages. Pathways enrichment was investigated using the WebGestalt software packages. KEGG and Pathway Commons databases were screened. Enrichment of DNA binding sites was also investigated though WebGestalt using the Transfac database. Bonferroni-adjusted P values of the hypergeometric test were used to determine enrichment significance. Geneset enrichment analysis was performed using GSEA v2.0.14 software with 1000 phenotype permutations100. Gene sets were downloaded from MSigDB101. Enrichment maps were generated with the Enrichment Map Plugin 1.3102 developed for Cytoscape 2.8103 using the default parameters.
Availability of data and materials
All data used in this work are available through the following databases: ONCOMINE (https://www.oncomine.org/), ATCG (https://cancergenome.nih.gov/), cBioportal (http://www.cbioportal.org/) and PrognoScan databases (http://www.abren.net/PrognoScan/).
Electronic supplementary material
Acknowledgements
We thank M. Yaniv, M. García-Domínguez and María Ceballos-Chávez for critical reading of the manuscript and helpful discussions. This work was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) [BFU2014-53543-P to J.C.R.], Junta de Andalucía (BIO-0321), and the European Union (FEDER). CABIMER is a Center partially funded by the Junta de Andalucía. The results published here are in part based on data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Author Contributions
J.A.G.-M. and J.C.R. contributed to experimental design, acquisition, analysis and interpretation of data. J.C.R. wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20217-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 2.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nature medicine. 2012;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 5.Wang W, et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 6.Euskirchen G, Auerbach RK, Snyder M. SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J Biol Chem. 2012;287:30897–30905. doi: 10.1074/jbc.R111.309302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. Embo J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 10.Vicent GP, et al. Two chromatin remodeling activities cooperate during activation of hormone responsive promoters. PLoS Genet. 2009;5:e1000567. doi: 10.1371/journal.pgen.1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King, H. W. & Klose, R. J. The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. eLife6, 10.7554/eLife.22631 (2017). [DOI] [PMC free article] [PubMed]
- 12.Shi J, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris SA, et al. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat Struct Mol Biol. 2014;21:73–81. doi: 10.1038/nsmb.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subtil-Rodriguez A, Reyes JC. BRG1 helps RNA polymerase II to overcome a nucleosomal barrier during elongation, in vivo. EMBO Rep. 2010;11:751–757. doi: 10.1038/embor.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Dieuleveult M, et al. Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature. 2016;530:113–116. doi: 10.1038/nature16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brownlee PM, Meisenberg C, Downs JA. The SWI/SNF chromatin remodelling complex: Its role in maintaining genome stability and preventing tumourigenesis. DNA Repair (Amst) 2015;32:127–133. doi: 10.1016/j.dnarep.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 17.Hodges, C., Kirkland, J. G. & Crabtree, G. R. The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer. Cold Spring Harb Perspect Med6, 10.1101/cshperspect.a026930 (2016). [DOI] [PMC free article] [PubMed]
- 18.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F, Delattre O. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol. 2015;10:145–171. doi: 10.1146/annurev-pathol-012414-040445. [DOI] [PubMed] [Google Scholar]
- 20.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS ONE. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helming KC, Wang X, Roberts CW. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer cell. 2014;26:309–317. doi: 10.1016/j.ccr.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson BG, et al. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Mol Cell Biol. 2014;34:1136–1144. doi: 10.1128/MCB.01372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutta A, et al. Composition and Function of Mutant Swi/Snf Complexes. Cell reports. 2017;18:2124–2134. doi: 10.1016/j.celrep.2017.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet. 2017;49:289–295. doi: 10.1038/ng.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jelinic P, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet. 2014;46:424–426. doi: 10.1038/ng.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witkowski L, et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet. 2014;46:438–443. doi: 10.1038/ng.2931. [DOI] [PubMed] [Google Scholar]
- 27.Ramos P, et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat Genet. 2014;46:427–429. doi: 10.1038/ng.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medina PP, et al. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Human mutation. 2008;29:617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 30.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dulak AM, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love C, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63:560–566. [PubMed] [Google Scholar]
- 34.Dunaief JL, et al. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 35.Wong AK, et al. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60:6171–6177. [PubMed] [Google Scholar]
- 36.Glaros S, Cirrincione GM, Palanca A, Metzger D, Reisman D. Targeted knockout of BRG1 potentiates lung cancer development. Cancer Res. 2008;68:3689–3696. doi: 10.1158/0008-5472.CAN-07-6652. [DOI] [PubMed] [Google Scholar]
- 37.Bultman S, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/S1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 38.Bultman SJ, et al. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27:460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- 39.Glaros S, et al. The reversible epigenetic silencing of BRM: implications for clinical targeted therapy. Oncogene. 2007;26:7058–7066. doi: 10.1038/sj.onc.1210514. [DOI] [PubMed] [Google Scholar]
- 40.Karnezis AN, et al. Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type. The Journal of pathology. 2016;238:389–400. doi: 10.1002/path.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reyes JC, et al. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) Embo J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muchardt C, Bourachot B, Reyes JC, Yaniv M. ras transformation is associated with decreased expression of the brm/SNF2alpha ATPase from the mammalian SWI-SNF complex. Embo J. 1998;17:223–231. doi: 10.1093/emboj/17.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reisman DN, et al. Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene. 2002;21:1196–1207. doi: 10.1038/sj.onc.1205188. [DOI] [PubMed] [Google Scholar]
- 44.Buscarlet M, et al. Essential role of BRG, the ATPase subunit of BAF chromatin remodeling complexes, in leukemia maintenance. Blood. 2014;123:1720–1728. doi: 10.1182/blood-2013-02-483495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Q, et al. The SWI/SNF ATPases Are Required for Triple Negative Breast Cancer Cell Proliferation. J Cell Physiol. 2015;230:2683–2694. doi: 10.1002/jcp.24991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jubierre L, et al. BRG1/SMARCA4 is essential for neuroblastoma cell viability through modulation of cell death and survival pathways. Oncogene. 2016;35:5179–5190. doi: 10.1038/onc.2016.50. [DOI] [PubMed] [Google Scholar]
- 47.Rhodes DR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia (New York, N.Y. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC medical genomics. 2009;2:18. doi: 10.1186/1755-8794-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cancer Genome Atlas Research Network. Electronic address, w. b. e. & Cancer Genome Atlas Research, N. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell169, 1327–1341 e1323, 10.1016/j.cell.2017.05.046 (2017). [DOI] [PMC free article] [PubMed]
- 51.Robertson AG, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder. Cancer. Cell. 2017;171:540–556 e525. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cancer Genome Atlas, N. Genomic Classification of Cutaneous Melanoma. Cell161, 1681–1696, 10.1016/j.cell.2015.05.044 (2015). [DOI] [PMC free article] [PubMed]
- 53.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature499, 43–49, 10.1038/nature12222 (2013). [DOI] [PMC free article] [PubMed]
- 54.Tronche, F. & Yaniv, M. Liver Gene Expression. (Austin: R. G. Landes Co, 1994).
- 55.Lin KB, et al. The Rap GTPases regulate the migration, invasiveness and in vivo dissemination of B-cell lymphomas. Oncogene. 2010;29:608–615. doi: 10.1038/onc.2009.345. [DOI] [PubMed] [Google Scholar]
- 56.Struckhoff AP, Rana MK, Worthylake RA. RhoA can lead the way in tumor cell invasion and metastasis. Front Biosci (Landmark Ed) 2011;16:1915–1926. doi: 10.2741/3830. [DOI] [PubMed] [Google Scholar]
- 57.Citterio C, et al. The rho exchange factors vav2 and vav3 control a lung metastasis-specific transcriptional program in breast cancer cells. Science signaling. 2012;5:ra71. doi: 10.1126/scisignal.2002962. [DOI] [PubMed] [Google Scholar]
- 58.Song C, et al. Expression of p114RhoGEF predicts lymph node metastasis and poor survival of squamous-cell lung carcinoma patients. Tumour Biol. 2013;34:1925–1933. doi: 10.1007/s13277-013-0737-8. [DOI] [PubMed] [Google Scholar]
- 59.Ura K, et al. Enhanced RASGEF1A expression is involved in the growth and migration of intrahepatic cholangiocarcinoma. Clin Cancer Res. 2006;12:6611–6616. doi: 10.1158/1078-0432.CCR-06-0783. [DOI] [PubMed] [Google Scholar]
- 60.Tong GX, et al. Expression of PAX8 in normal and neoplastic renal tissues: an immunohistochemical study. Mod Pathol. 2009;22:1218–1227. doi: 10.1038/modpathol.2009.88. [DOI] [PubMed] [Google Scholar]
- 61.Barr ML, et al. PAX-8 expression in renal tumours and distant sites: a useful marker of primary and metastatic renal cell carcinoma? J Clin Pathol. 2015;68:12–17. doi: 10.1136/jclinpath-2014-202259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barras D, Widmann C. GAP-independent functions of DLC1 in metastasis. Cancer Metastasis Rev. 2014;33:87–100. doi: 10.1007/s10555-013-9458-0. [DOI] [PubMed] [Google Scholar]
- 63.Tripathi, B. K. & Lowy, D. R. DLC1: a tumor suppressor that regulates Rho signaling. Oncotarget, 10.18632/oncotarget.16805 (2017). [DOI] [PMC free article] [PubMed]
- 64.Ratner N, Miller SJ. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 2015;15:290–301. doi: 10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schell C, Wanner N, Huber TB. Glomerular development–shaping the multi-cellular filtration unit. Semin Cell Dev Biol. 2014;36:39–49. doi: 10.1016/j.semcdb.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 66.Brunskill EW, Potter SS. RNA-Seq defines novel genes, RNA processing patterns and enhancer maps for the early stages of nephrogenesis: Hox supergenes. Dev Biol. 2012;368:4–17. doi: 10.1016/j.ydbio.2012.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu, Y., Ye, Y., Yang, Q. & Shi, S. Single-cell RNA-sequence analysis of mouse glomerular mesangial cells uncovers mesangial cell essential genes. Kidney Int, 10.1016/j.kint.2017.01.016 (2017). [DOI] [PubMed]
- 68.Scarpulla RC. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann N Y Acad Sci. 2008;1147:321–334. doi: 10.1196/annals.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wallace DC. Mitochondria and cancer: Warburg addressed. Cold Spring Harb Symp Quant Biol. 2005;70:363–374. doi: 10.1101/sqb.2005.70.035. [DOI] [PubMed] [Google Scholar]
- 70.Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–246. doi: 10.1016/S0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 71.Jiang W, et al. Intratumoral heterogeneity analysis reveals hidden associations between protein expression losses and patient survival in clear cell renal cell carcinoma. Oncotarget. 2017;8:37423–37434. doi: 10.18632/oncotarget.16965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin H, Wong RP, Martinka M, Li G. BRG1 expression is increased in human cutaneous melanoma. Br J Dermatol. 2010;163:502–510. doi: 10.1111/j.1365-2133.2010.09851.x. [DOI] [PubMed] [Google Scholar]
- 73.Saladi SV, et al. Modulation of extracellular matrix/adhesion molecule expression by BRG1 is associated with increased melanoma invasiveness. Molecular cancer. 2010;9:280. doi: 10.1186/1476-4598-9-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu Q, et al. Targeting the chromatin remodeling enzyme BRG1 increases the efficacy of chemotherapy drugs in breast cancer cells. Oncotarget. 2016;7:27158–27175. doi: 10.18632/oncotarget.8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bai J, et al. BRG1 is a prognostic marker and potential therapeutic target in human breast cancer. PLoS ONE. 2013;8:e59772. doi: 10.1371/journal.pone.0059772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Do SI, et al. Increased Brahma-related Gene 1 Expression Predicts Distant Metastasis and Shorter Survival in Patients with Invasive Ductal Carcinoma of the Breast. Anticancer Res. 2016;36:4873–4882. doi: 10.21873/anticanres.11051. [DOI] [PubMed] [Google Scholar]
- 77.Kaufmann B, et al. BRG1 promotes hepatocarcinogenesis by regulating proliferation and invasiveness. PLoS ONE. 2017;12:e0180225. doi: 10.1371/journal.pone.0180225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu Q, et al. The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer. Epigenomics. 2017;9:919–931. doi: 10.2217/epi-2017-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stepanenko AA, Vassetzky YS, Kavsan VM. Antagonistic functional duality of cancer genes. Gene. 2013;529:199–207. doi: 10.1016/j.gene.2013.07.047. [DOI] [PubMed] [Google Scholar]
- 80.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 81.LeGouy E, Thompson EM, Muchardt C, Renard JP. Differential preimplantation regulation of two mouse homologues of the yeast SWI2 protein. Dev Dyn. 1998;212:38–48. doi: 10.1002/(SICI)1097-0177(199805)212:1<38::AID-AJA4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 82.Nejak-Bowen KN, Monga SP. Beta-catenin signaling, liver regeneration and hepatocellular cancer: sorting the good from the bad. Semin Cancer Biol. 2011;21:44–58. doi: 10.1016/j.semcancer.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huch M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nagl NG, Jr, Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. Embo J. 2007;26:752–763. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Asp P, Wihlborg M, Karlen M, Farrants AK. Expression of BRG1, a human SWI/SNF component, affects the organisation of actin filaments through the RhoA signalling pathway. J Cell Sci. 2002;115:2735–2746. doi: 10.1242/jcs.115.13.2735. [DOI] [PubMed] [Google Scholar]
- 87.Tuveson DA, Jacks T. Modeling human lung cancer in mice: similarities and shortcomings. Oncogene. 1999;18:5318–5324. doi: 10.1038/sj.onc.1203107. [DOI] [PubMed] [Google Scholar]
- 88.Damiano L, et al. Oncogenic targeting of BRM drives malignancy through C/EBPbeta-dependent induction of alpha5 integrin. Oncogene. 2014;33:2441–2453. doi: 10.1038/onc.2013.220. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89.Albini S, et al. Brahma is required for cell cycle arrest and late muscle gene expression during skeletal myogenesis. EMBO Rep. 2015;16:1037–1050. doi: 10.15252/embr.201540159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Endo M, et al. Alterations of the SWI/SNF chromatin remodelling subunit-BRG1 and BRM in hepatocellular carcinoma. Liver Int. 2013;33:105–117. doi: 10.1111/liv.12005. [DOI] [PubMed] [Google Scholar]
- 91.Jiang W, et al. Immunohistochemistry Successfully Uncovers Intratumoral Heterogeneity and Widespread Co-Losses of Chromatin Regulators in Clear Cell Renal Cell Carcinoma. PLoS ONE. 2016;11:e0164554. doi: 10.1371/journal.pone.0164554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reisman DN, Sciarrotta J, Bouldin TW, Weissman BE, Funkhouser WK. The expression of the SWI/SNF ATPase subunits BRG1 and BRM in normal human tissues. Appl Immunohistochem Mol Morphol. 2005;13:66–74. doi: 10.1097/00129039-200503000-00011. [DOI] [PubMed] [Google Scholar]
- 93.Wang X, et al. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res. 2009;69:8094–8101. doi: 10.1158/0008-5472.CAN-09-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.St Pierre R, Kadoch C. Mammalian SWI/SNF complexes in cancer: emerging therapeutic opportunities. Curr Opin Genet Dev. 2017;42:56–67. doi: 10.1016/j.gde.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cox DR. Regression models and life tables. J Roy Statist Soc B. 1972;34:187–220. [Google Scholar]
- 96.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 97.t Hoen PA, et al. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 99.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liberzon A, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Okayama H, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 105.Pawitan Y, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gobble RM, et al. Expression profiling of liposarcoma yields a multigene predictor of patient outcome and identifies genes that contribute to liposarcomagenesis. Cancer Res. 2011;71:2697–2705. doi: 10.1158/0008-5472.CAN-10-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this work are available through the following databases: ONCOMINE (https://www.oncomine.org/), ATCG (https://cancergenome.nih.gov/), cBioportal (http://www.cbioportal.org/) and PrognoScan databases (http://www.abren.net/PrognoScan/).