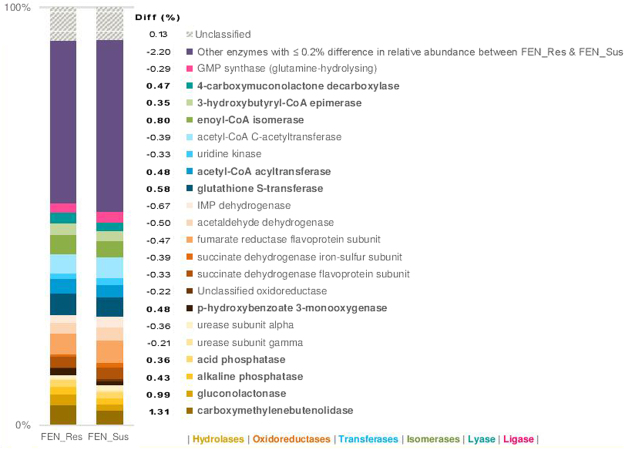
Figure 4.
Putative bacterial enzymes involved in microbial xenobiotic degradation in fenitrothion resistant and susceptible An. albimanus. The plot shows the proportion (%) of sequencing reads (FEN_Res, n = 458,614; and FEN_Sus, n = 311,919) aligned to bacterial enzymes in the microbial xenobiotic degradation pathway, and the difference in relative abundance of identified enzymes between fenitrothion resistant and susceptible An. albimanus (Diff.). Only enzymes with Diff. ≥ 0.2% at p < 0.0001 are shown, and significantly enriched enzymes in FEN_Res are presented in bold typeface.