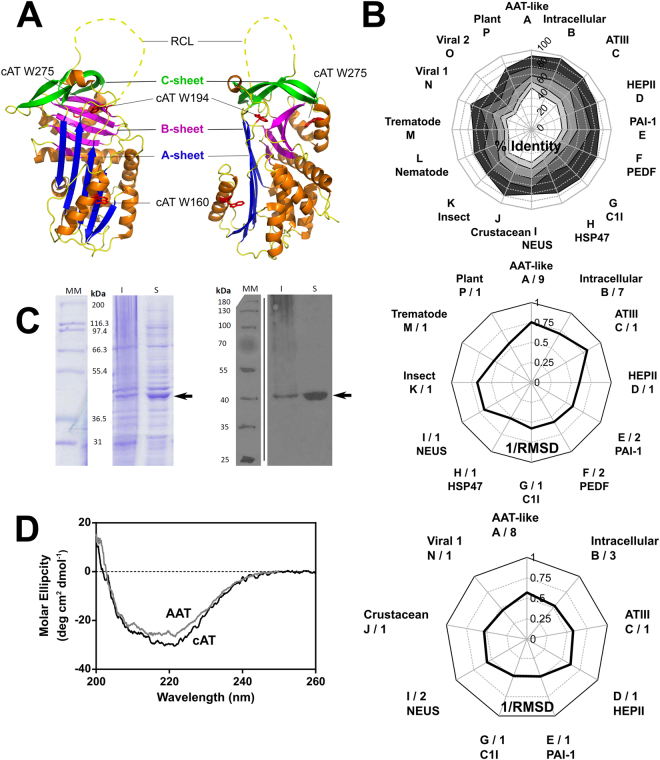
Figure 1.
Properties of a conserpin variant. (A) The serpin fold is comprised of three β-sheets and eight or nine α-helices, illustrated using a cartoon representation of native conserpin (PDB 5CDX)34. Connecting β-strand 5 A and β-strand 1 C is the solvent exposed reactive centre loop (RCL) which dictates the target protease specificity of inhibitory serpins (unresolved in the structure and therefore denoted by a dashed line). Upon proteolytic cleavage, the RCL becomes a 6th central strand of the normally 5-stranded β-sheet A. The positions of the tryptophan residues mutated in this study are indicated. (B) Upper panel: Radial plot of the average sequence identity between the consensus sequence and members of individual serpin phylogenetic clades1, calculated using all alignment positions (white central area), or the 75%, 50%, 25% and 10% lowest variability sites (shaded from light grey to dark grey). Middle panel: Reciprocal RMSD of native conserpin (PDB 5CDX) compared with the common core of representative structures from different phylogenetic clades. Lower panel: A comparison between latent conserpin (PDB 5CDZ) and representative cleaved/latent structures. (C) Left: SDS-PAGE (10% w/v, see full gels in Supplementary Fig. S1) section of cAT after 20 hours of auto-induction, visualised using Coomassie Blue stain. Right: Western blot of the corresponding SDS-PAGE, developed using an anti-His6 antibody. Lane MM, molecular weight markers; I, insoluble fraction; S, soluble lysate fraction. cAT is indicated by the arrow. (D) Far-UV spectrum of cAT (black) compared with α1-AT (grey) at a concentration of 0.2 mg mL−1, using a path length of 0.1 cm.