Figure 1.
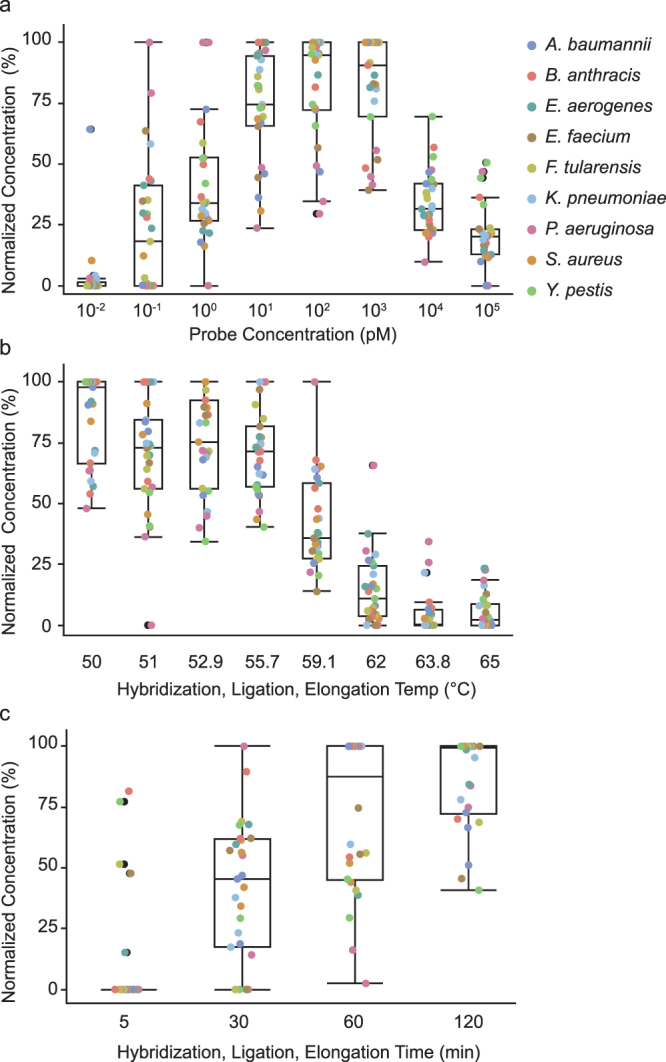
MIP protocol optimization for the 16S probeset. Pooled 16S MIPs were tested against DNA extracts of representative biothreats and ESKAPE pathogens to optimize target capture conditions including (a) probe concentration, (b) reaction temperature and (c) reaction duration. Amplicon formation was quantitatively measured after probe circularization and amplification with the universal primer set across three replicates for each organism at each variable. Concentrations were normalized to 0% and 100% representing the lowest and highest concentrations across the experimental range. Data points for each organism were combined and a corresponding outlier box plot was generated for each variable.
