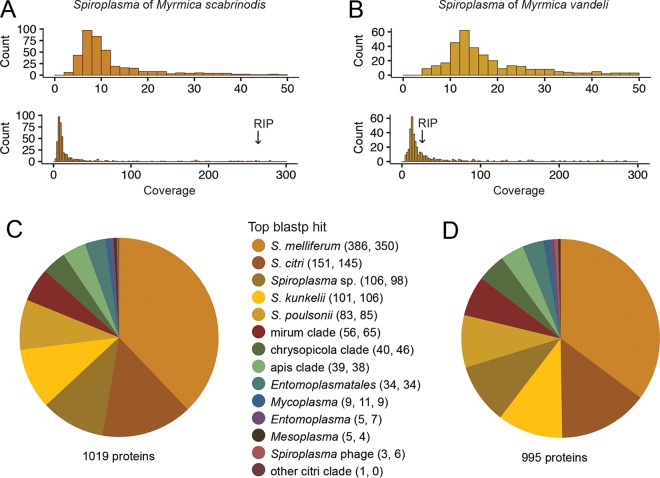
FIG 3.
Genome coverage and Spiroplasma gene assignment of Myrmica symbionts. Read coverage distribution graphed in a 0 to 50 range and 0 to 300 range for M. scabrinodis (A) and M. vandeli (B) Spiroplasma symbionts. Read coverage of the RIP-encoding contig for each symbiont is indicated with arrows. Pie charts summarize gene assignments within each symbiont's genome. The majority fraction of Spiroplasma genes in M. scabrinodis (C) and M. vandeli (D) Spiroplasma symbionts match best by blastp to the honeybee pathogen Spiroplasma melliferum, and overall, most genes match to members of the citri clade: Spiroplasma citri, S. kunkelii, S. melliferum, and S. poulsonii. The respective numbers of top blastp matches from M. scabrinodis and M. vandeli symbionts are indicated in parentheses.