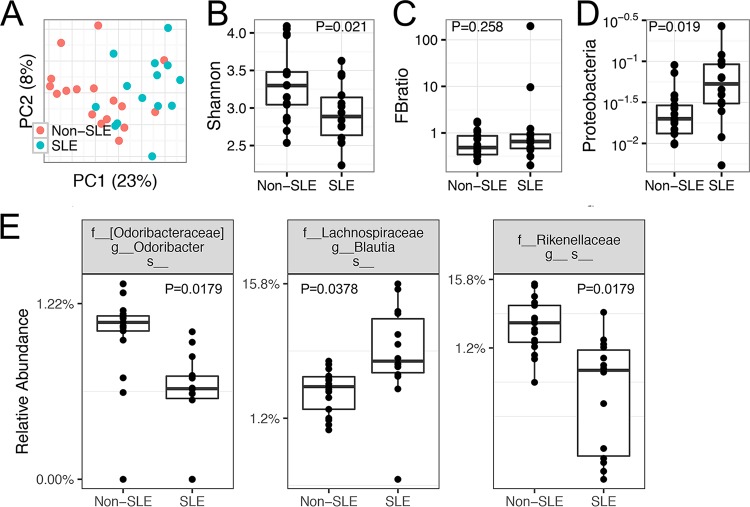
FIG 3.
Gut microbiota dysbiosis in SLE patients with active disease, compared to non-SLE controls. The inclusion and exclusion criteria are listed in Materials and Methods. A fecal sample was collected from each patient or control subject at the time of the first visit, and the samples were subjected to 16S rRNA sequencing analysis. (A) PCoA plot, showing no separation of overall bacterial structures (P > 0.05). (B) Significant difference in bacterial diversity levels, as indicated by the Shannon index (P < 0.05). (C) No significant difference in Firmicutes/Bacteroidetes (FB) ratios (P > 0.05). (D) Significant difference in the relative abundance of the phylum Proteobacteria, a representation of facultative anaerobic Gram-negative bacteria (P < 0.05). (E) Significant changes in several bacterial species, with the P values indicated (nonparametric Mann-Whitney test). DESeq2 analysis was also performed, and the DESeq2 P values were 0.037, 0.007, and 0.007, respectively. P values were corrected for multiple comparisons by controlling the FDRs.