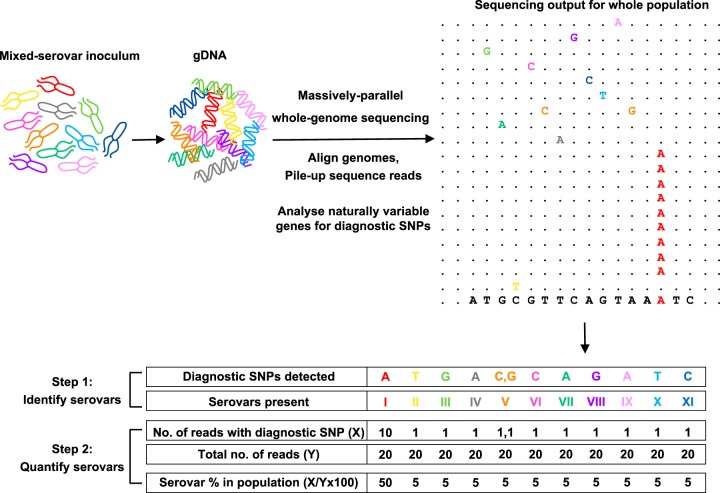
FIG 1.
Approach to quantify bacterial strains in a mixed-strain population. Whole-genome sequencing followed by the analysis of known naturally variable genes for strain-specific SNPs can determine the composition of a mixed-strain population. S. enterica serovars can be distinguished by serovar-specific diagnostic SNPs in rpoB and ileS. When a mixed-serovar population is sequenced and aligned to a reference genome, these diagnostic SNPs can be used to identify the serovars in the population (step 1), and from the number of reads with diagnostic SNPs, the abundance of each serovar can be quantified (step 2). For example, here, the diagnostic SNP for serovar I (A) is detected 10 times out of a total 20 reads. Thus, it makes up 50% of the population. Diagnostic SNPs for the other serovars are detected only once. Thus, they each make up 5% of the population. When more than one diagnostic SNP is present, as seen for serovar V (C, G), the average abundance (5% at C and 5% at G) is used to estimate the overall abundance of that serovar in the population.