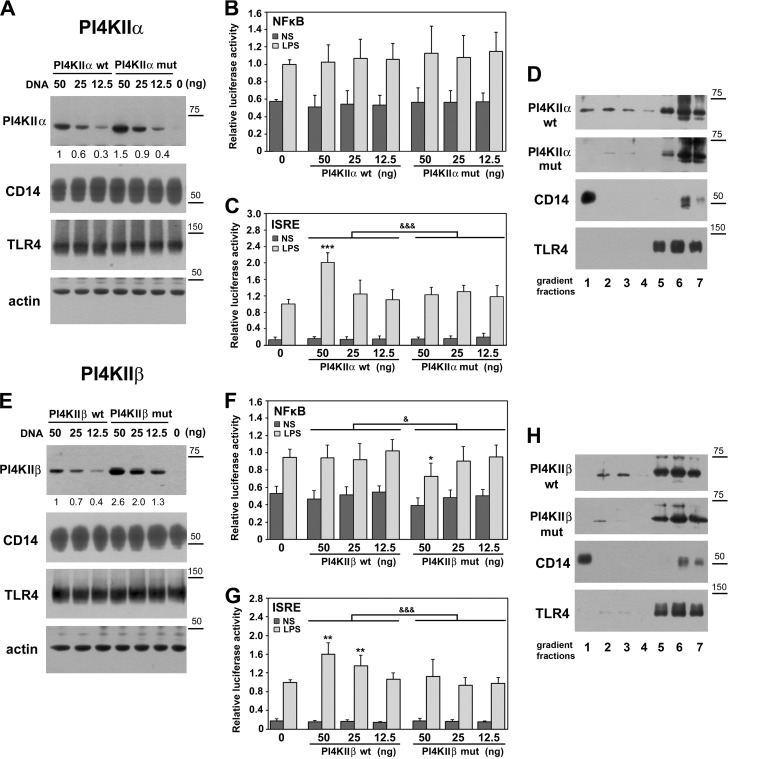
Fig. 6.
PI4KIIα and PI4KIIβ up-regulate activity of IRF(s) in palmitoylation-dependent manner. Palmitoylation affects subcellular distribution of kinases. HEK293 cells were co-transfected with plasmids encoding CD14 and FLAG-tagged TLR4/MD2 and indicated amounts of plasmids encoding Myc-tagged PI4KIIα (A–D) or PI4KIIβ (E–H) kinases wild type (wt) or their deletion mutants lacking the S-palmitoylation site (mut). In control cells, PI4KII-bearing plasmids were omitted (“0”). In (A–C) and (E–G) the DNA mixture used for cell transfection also contained plasmid encoding Renilla luciferase and firefly luciferase controlled by either NFκB-dependent or ISRE-dependent promoter. Cells were either left unstimulated (NS) or were stimulated with 100 ng/ml LPS for 24 h. (A, E) Immunoblotting analysis of ectopically expressed PI4KIIα, PI4KIIβ, CD14, and TLR4 using anti-Myc, anti-CD14 or anti-FLAG antibodies. Equal protein loading was verified by probing membranes with anti-actin antibody. Numbers below upper panels indicate relative mean level of PI4KIIα or PI4KIIβ quantified by densitometry in four experiments. (B, F) NFκB activity and (C, G) ISRE activity assessed by corresponding firefly luciferase activity measured in cell lysates. The firefly luciferase activity was normalized against constitutive Renilla luciferase activity in the same sample. The relative luciferase activity (RLA) is expressed as a fold increase over the RLA value found in LPS-stimulated cells lacking the ectopically expressed PI4KIIα or PI4KIIβ (“0”). Data are mean ± s.d. from four experiments. &, &&&, Significant differences at p < 0.05 and p < 0.001 between LPS-stimulated samples established with two-way ANOVA with interactions. *, **, ***, Significantly different at p < 0.05, p < 0.01, and p < 0.001 from RLA in LPS-stimulated cells lacking the ectopically expressed PI4KIIα or PI4KIIβ (“0”), as estimated using 1-way ANOVA with Tukey's post hoc test. (D, H) Distribution of PI4KIIα (D) and PI4KIIβ (H) in density gradient fractions. HEK293 transfectants were lysed in 0.5% Brij 98 and the lysates were fractionated over 0–40% OptiPrep density gradient. Seven fractions with increasing density were collected and analyzed for the presence of indicated proteins. Molecular weight markers are shown on the right.