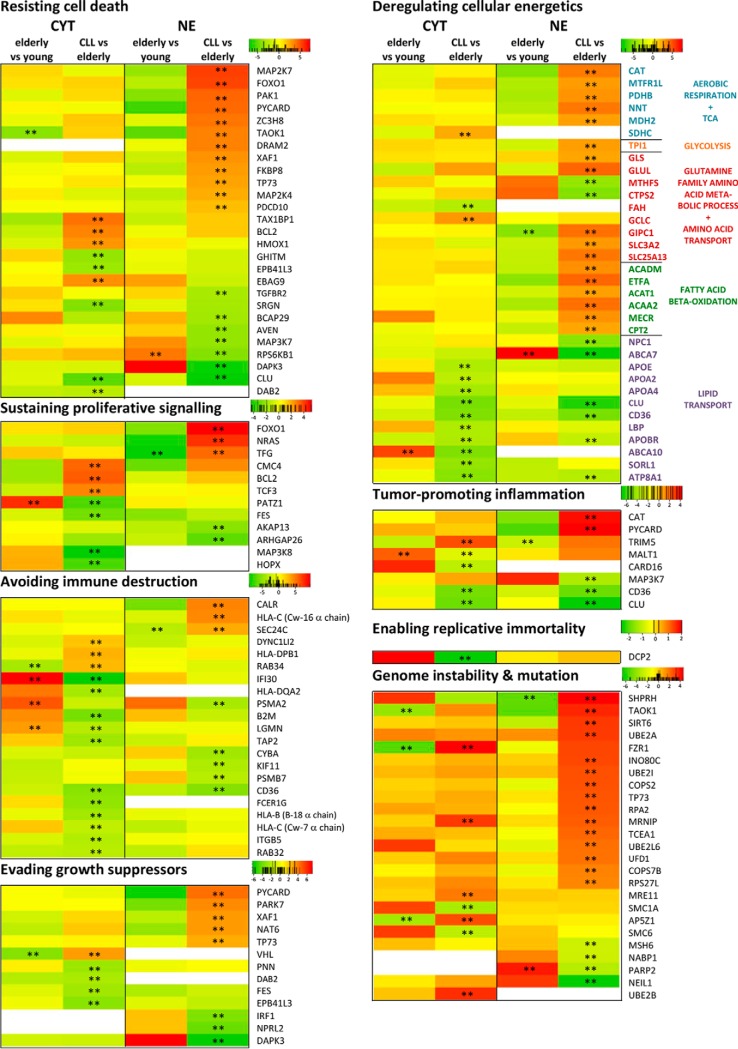
Fig. 3.
Heat maps for proteins related to cancer hallmarks (54) relevant for CLL. Heat maps are shown for proteins that were significantly regulated (FDR<0.05, indicated by **) in at least one fraction between CLL samples and samples from age-matched healthy donors. Red color indicates up-regulation, green color down-regulation, and white color absence of the protein in the respective fraction. Numbers given in the color scales represent fold changes in the form of 2x. Proteins were associated with the respective cancer hallmark term based on Uniprot GO terminology and/or keywords. So, to assign proteins to the hallmarks “Resisting cell death,” “Sustained proliferative signaling,” and “Evading growth suppression,” the Uniprot keywords “Apoptosis,” “Proto-oncogene,” and “Tumor suppressor” were utilized, respectively. To associate proteins to the hallmark “Deregulating cellular energetics,” a combined list of the GO terms “Aerobic respiration,” “Glycolysis,” “Fatty acid beta-oxidation,” “TCA cycle,” “Glutamine family amino acid metabolic process,” “Lipid transport,” “Glucose transport,” and “Amino acid transport” was used. Proteins involved in “Avoiding immune destruction” were determined using the GO terms “Antigen processing and presentation.” The Uniprot GO terms “positive regulation of NF-kappaB transcription factor activity,” “DNA repair,” and “negative regulation of telomere maintenance via telomerase” were utilized for the cancer hallmarks “Tumor-promoting inflammation,” “Genome instability and mutation,” and “Enabling replicative immortality,” respectively.