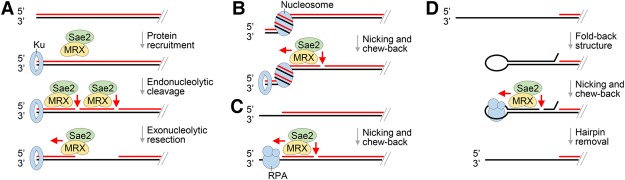
Figure 4.
Models for MRX action in DNA end resection and processing of protein obstacles. (A) In DSB end resection, Ku, while protecting the DNA end from exonucleolytic digestion, promotes 5′ strand incision by MRX–Sae2. The resulting DNA nick is enlarged exonucleolytically in the 3′–5′ direction by MRX–Sae2. (B) Within the context of chromatin, MRX–Sae2 endonucleolytically cleaves the 5′ strand of DNA that borders a nucleosome. The resulting DNA nick is expected to be subject to 3′–5′ chew-back to generate a DNA gap. (C) Binding of RPA to a partially resected DNA end activates 5′ strand scission by MRX–Sae2, which would lead to a gap upon 3′–5′ chew-back. (D) The fold-back structure, which arises from annealing between inverted repeats in ssDNA, is removed by MRX–Sae2 in collaboration with RPA via nicking and chew-back action.