Figure 6.
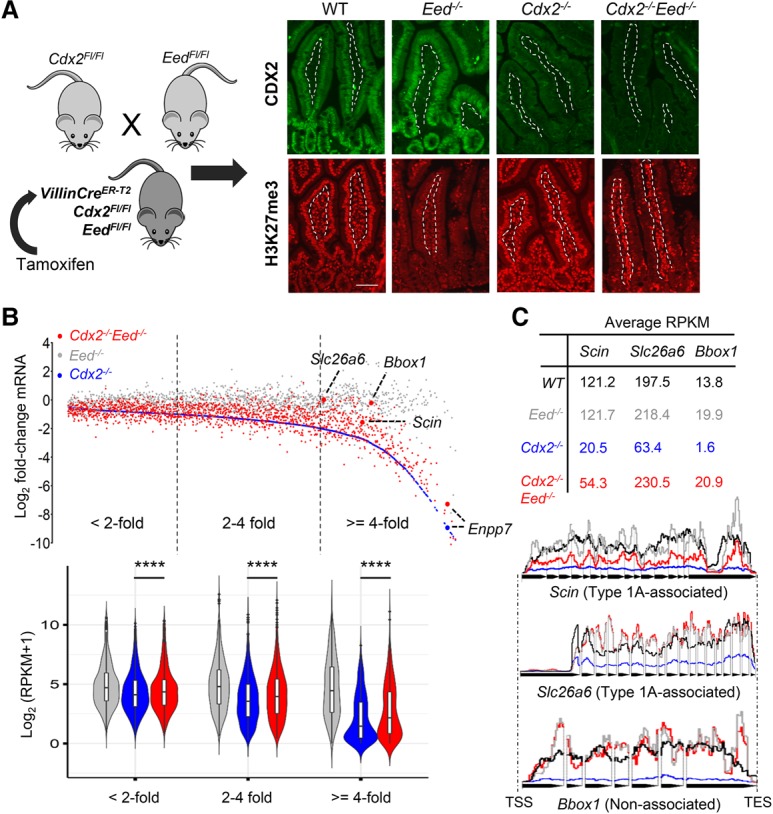
Deletion of the PRC2 component gene Eed eliminates H3K27me3 and partially rescues CDX2-dependent gene expression. (A) Mouse breeding and treatment scheme to eliminate both CDX2 and PRC2 activity, verified by CDX2 (top) and H3K27me3 (bottom) immunostains on the third day after 5 d of tamoxifen treatment to activate Cre. Dashed lines demarcate the epithelium, selectively targeted by Villin-Cre, from the intestinal lamina propria. Bar, 10 µm. (B) Scatter plot of all genes reduced in Cdx2−/− (blue; arranged in order of increasing effect) and their levels in Eed−/− (gray) and Cdx2−/−;Eed−/− (red) intestinal cells. Dots represent the mean from two replicates of each genotype, and the violin plots below represent RPKM values of bins of genes that decrease less than twofold, twofold to fourfold, or fourfold or greater in Cdx2−/− villi. (****) P < 0.0001. Boxes within the violins show median values at the 75th and 25th percentiles, and the whiskers show 1.5 times the interquartile range. Significance was determined by the Friedman test. EED loss substantially rescued many transcripts, especially among those most affected in Cdx2−/− cells, with some genes restored to wild-type levels. (C) Slc26a6 and Scin (two type 1A-associated genes) and Bbox1 (an unlinked gene) are highlighted by RPKM values and collated tracks (data were averaged from duplicates, introns excluded). In contrast, Enpp7 in B showed minimal rescue.
