Figure 2.
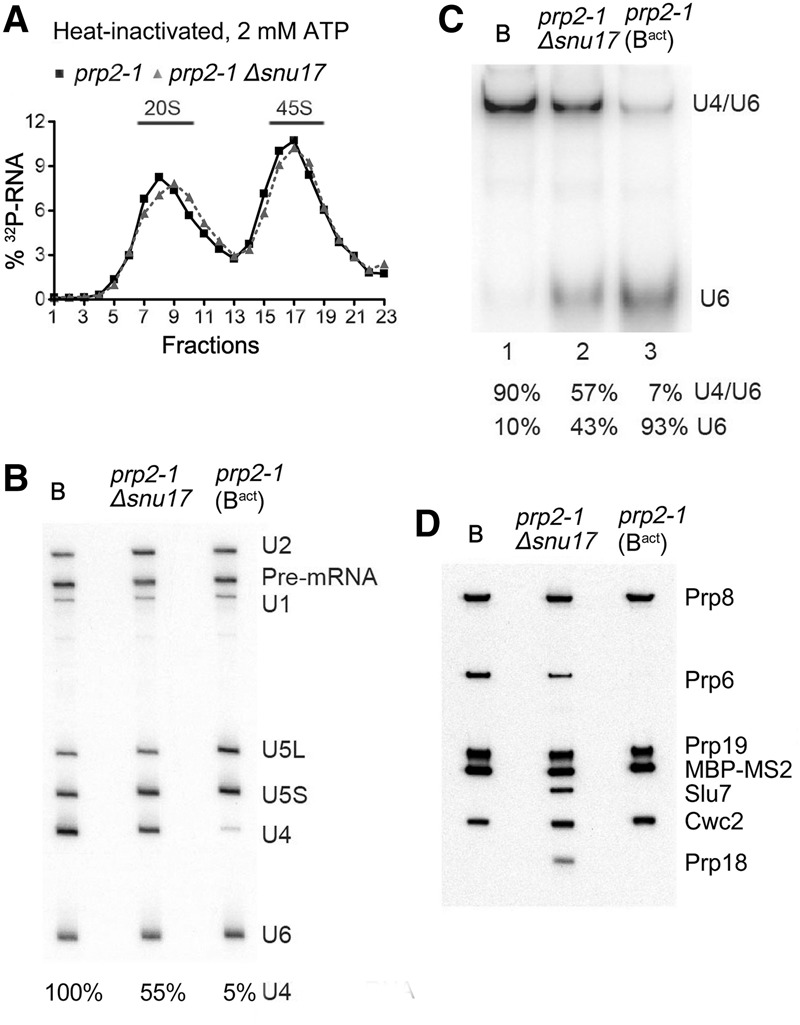
The transformation of the spliceosomal B complex into Bact is less efficient in the absence of RES complex proteins. (A) Glycerol gradient sedimentation profile of MS2-MBP affinity-purified spliceosomal complexes formed after initial heat inactivation of Prp2 on 32P-labeled Actin pre-mRNA in extracts from prp2-1 or prp2-1 Δsnu17 yeast cells after 60 min in the presence of 2 mM ATP. (B) RNA compositions of affinity-purified B complexes (formed at 50 µM ATP) or Bact or splicing complexes formed in extracts from prp2-1 Δsnu17 cells (at 2 mM ATP) sedimenting in the 45S peak fractions of the glycerol gradient. The pre-mRNA and snRNAs were separated by denaturing polyacrylamide gel electrophoresis (PAGE) and detected as described in the legend for Figure 1. The amount of U4 snRNA in each lane (shown below) was quantitated as a percentage of the pre-mRNA band (which acts as a spliceosome loading control) and then normalized to the amount of U4 in the B complex, which was set to 100%. (C) Quantitation of the amount of U4/U6 duplex and U6 snRNA in affinity-purified B, Bact, or spliceosomal complexes formed in extracts from prp2-1 Δsnu17 cells. RNAs were analyzed by native PAGE, and U6 snRNA was visualized by Northern blotting. The percent of free U6 versus U6 base-paired with U4 was quantitated with a PhosphorImager and is shown below each lane. The identity of the slower-migrating band as U4/U6 was confirmed by Northern blotting with a probe against U4 snRNA. (D) Proteins from affinity-purified B complexes (formed at 50 µM ATP) or spliceosomal complexes formed at 2 mM ATP in extracts from prp2-1 (predominantly Bact complexes) or prp2-1 Δsnu17 cells (as indicated) after heat inactivation of Prp2 were analyzed by Western blotting using antibodies against the indicated proteins. Antibodies against Prp8 were used to ensure equal loading.
