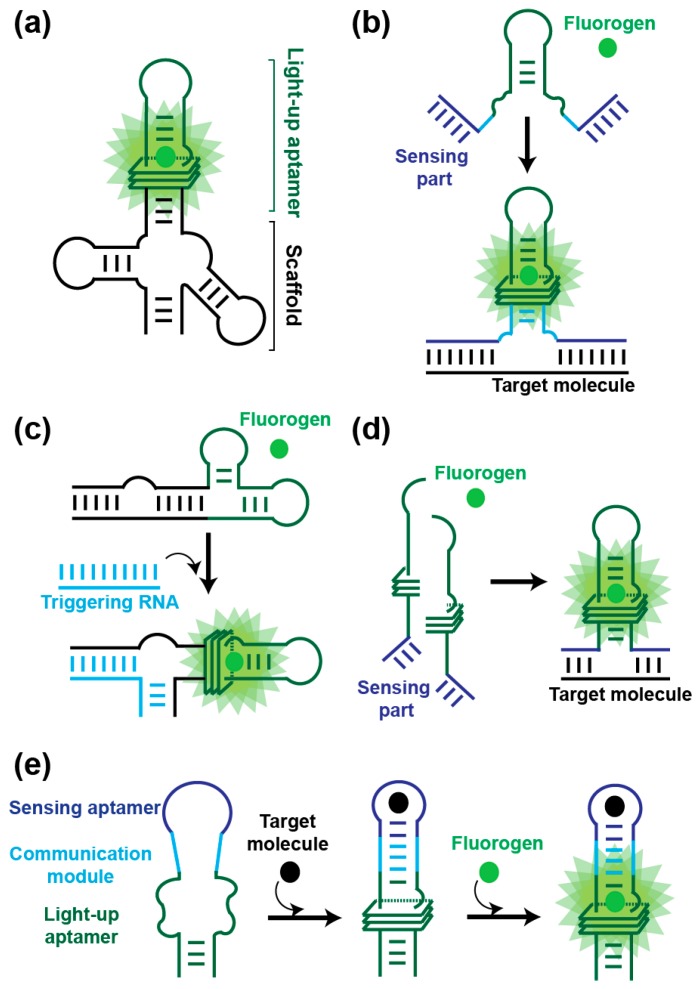
Figure 4.
Light-up aptamers engineering and uses thereof. (a) Light-up RNA aptamers can be embedded into a scaffold RNA (here a tRNA) to improve folding efficiency and resistance to RNases [29,61]. The construct can either be directly expressed from an independent promoter or inserted into the sequence of a target mRNA. (b) Light-up aptamer sensing a target RNA in trans [77,78]. A key helix (light blue) of the light-up aptamer is engineered to weaken the structure of the RNA and destabilized the fluorogen-binding site. Furthermore, sequences complementary to the target nucleic acid (acting as a sensing part) are appended to both ends of the molecule (dark blue). Upon binding to the target molecule (in black), the structure of the light-up moiety is stabilized and its fluorogen-binding capacity is restored, leading to fluorescence emission (green shadow). (c) Light-up aptamer sensing a target RNA in trans via strand displacement [79,80,81]. Sequences are appended to both ends of the light-up aptamer and induce an alternative folding preventing the interaction of the aptamer with the fluorogen. However, the binding of the target RNA to the construct induces a conformational change restoring the structure of the light-up aptamer moiety and its fluorogen-binding capacity, leading to fluorescence emission (green shadow). (d) Sensing target nucleic acids using split light-up aptamers [82,83]. The aptamer is split into two halves, each possessing a sequence complementary to the target nucleic acid (acting as a sensing part, in dark blue). Whereas both halves cannot form a functional aptamer in the absence of the target nucleic acid, the presence of the latter drives the productive association of both molecules into a functional light-up aptamer able to emit fluorescence. (e) Allosteric light-up RNA aptamer [84,85]. The light-up aptamer is engineered to weaken the structure of the RNA and destabilize the fluorogen-binding site. Moreover, the sequence of a sensing aptamer (in dark blue) is inserted into that of the light-up aptamer via a communication module (in light blue). Upon the interaction of the sensing aptamer with a target molecule (e.g., a metabolite or a protein), the structure of the light-up aptamer is stabilized and its fluorogen-binding capacity is restored. Note that all the examples are shown with the Spinach aptamer, but these concepts can be extended to any other light-up aptamer.