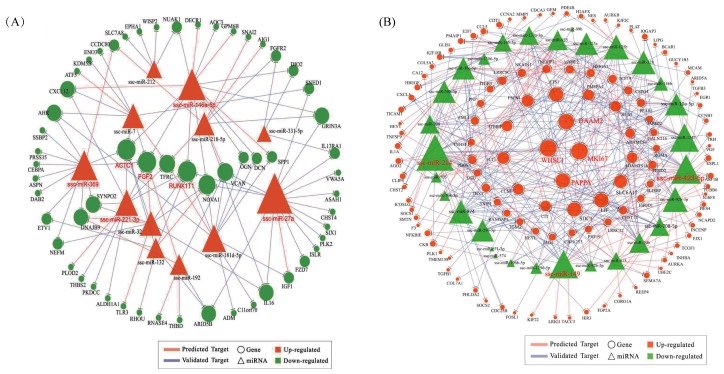
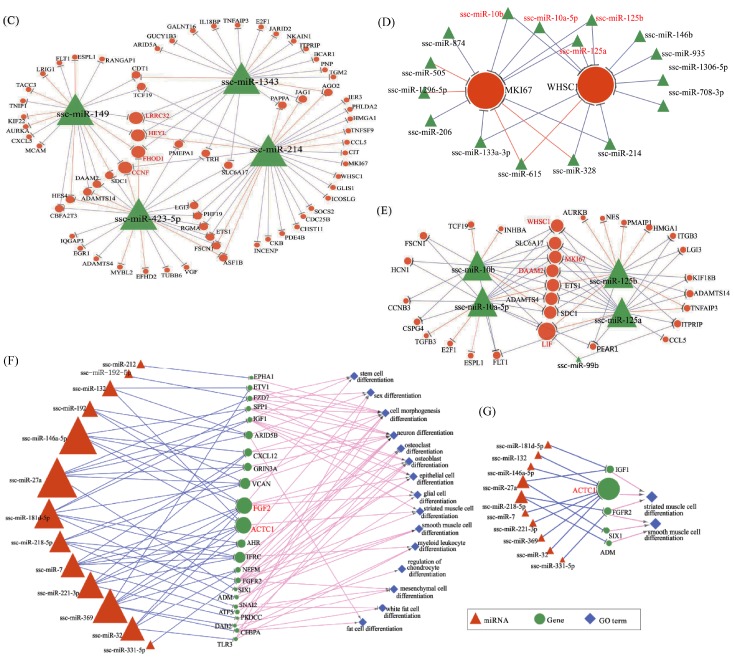
Figure 5.
Analyses of mRNA-miRNA networks in P-N1ICD vs. P-Control. (A) The network of upregulated miRNAs and downregulated genes in P-N1ICD vs. P-Control. Deep orange triangle represents upregulated miRNA; green circle represents downregulated gene; blue lines represent predicted targeting genes; orange lines represent verification of targeting genes. (B) The network of downregulated miRNAs and upregulated genes in proliferation. Green triangle represents downregulated miRNA; deep orange circle represents upregulated gene. (C) The network of four hub downregulated miRNAs (ssc-miR-214, ssc-miR-423-5p, ssc-miR-149 and ssc-miR-1343) in P-N1ICD vs. P-Control, the hub genes of which are leucine rich repeat containing 32 (LRRC32), hes related family bHLH transcription factor with YRPW motif-like (HEYL), formin homology 2 domain containing 1 (FHOD1) and cyclin F (CCNF). (D) The network of two hub upregulated genes (MKI67 and WHSC1). Both MKI67 and WHSC1 are targeted by ssc-miR-10b, ssc-miR-10a-5p, ssc-miR-125a and ssc-miR-125b, which belong to the miR-10 family. (E) The network of five downregulated miRNAs in proliferation, which belong to the miR-10 family. Interleukin 6 family cytokine (LIF) is targeted by all five miRNAs. Three hub genes in the Figure 4B network, including MKI67, WHSC1 and dishevelled associated activator of morphogenesis 2 (DAAM2), are targeted by ssc-miR-10b, ssc-miR-10a-5p, ssc-miR-125a and ssc-miR-125b. (F) The network of downregulated miRNAs, upregulated genes and differentiation-related GO terms. Blue rhombus represents differentiation-related GO terms. (G) The network of downregulated miRNAs, upregulated genes and muscle cell differentiation-related GO terms. Blue rhombus represents differentiation-related GO terms.