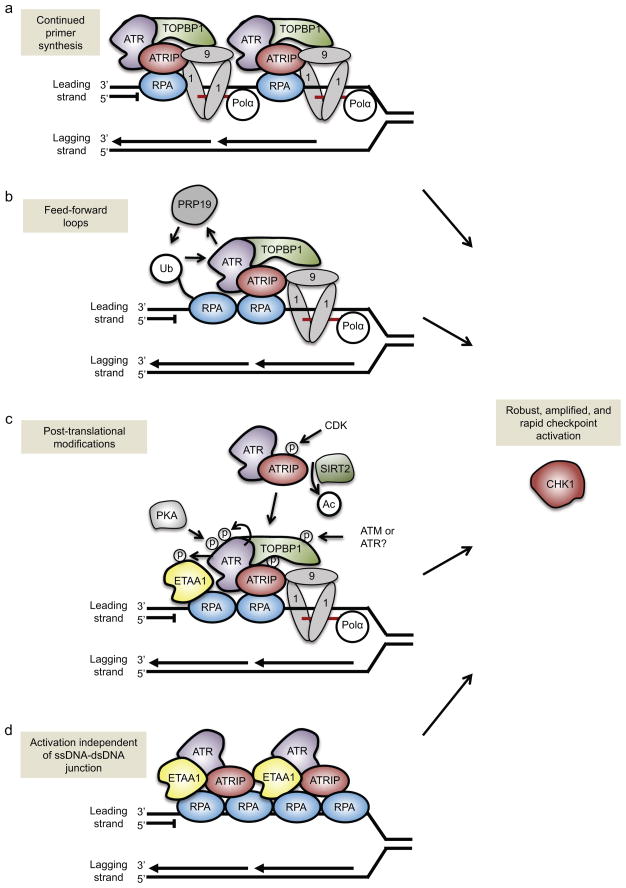
Figure 3. Amplification of ATR signaling.
There are multiple ways to amplify ATR signaling at individual replication forks. (a) Continued primer synthesis by DNA polymerase alpha (Pol α) ahead of the stalled leading-strand polymerase can generate multiple ssDNA–dsDNA junctions at a single fork. This would create multiple loading points for the RAD9–RAD1–HUS1 (9-1-1) complex and the ATR activator topoisomerase II binding protein 1 (TOPBP1), and, accordingly, would increase the number of ATR proteins that are activated at a single fork. (b) The E3 ubiquitin ligase PRP19 creates a feed-forward loop to amplify ATR activity. PRP19 ubiquitylates replication protein A (RPA), which increases ATR activity, which in turn boosts PRP19-dependent ubiquitylation of RPA. (c) Multiple post-translational modifications amplify ATR activation. These include protein kinase A (PKA) phosphorylation of ATR, ATR autophosphorylation, cyclin-dependent kinase 2 (CDK2) phosphorylation of ATR-interacting protein (ATRIP), ATR and/or ATM phosphorylation of TOPBP1, and potentially ATR phosphorylation of Ewing tumor-associated antigen 1 (ETAA1). ATRIP deacetylation by Sirtuin 2 (SIRT2) helps recruit ATR–ATRIP to the stalled replication fork. (d) The second ATR activator, ETAA1, can stimulate ATR activity independently of the presence of ssDNA–dsDNA junctions. Because ETAA1, ATR and ATRIP are recruited to RPA–ssDNA, longer stretches of ssDNA could recruit multiple ETAA1, ATR and ATRIP proteins to a single fork and produce an amplified ATR-mediated response.
CHK1, checkpoint kinase 1