Abstract
In this study, following its isolation from contaminated soil, the genomic sequence of Pseudomonas alcaliphila strain JAB1 (=DSM 26533), a biphenyl-degrading bacterium, is reported and analyzed in relation to its extensive degradative capabilities. The P. alcaliphila JAB1 genome (GenBank accession no. CP016162) consists of a single 5.34 Mbp-long chromosome with a GC content of 62.5%. Gene function was assigned to 3816 of the 4908 predicted genes. The genome harbors a bph gene cluster, permitting degradation of biphenyl and many congeners of polychlorinated biphenyls (PCBs), a ben gene cluster, enabling benzoate and its derivatives to be degraded, and phe gene cluster, which permits phenol degradation. In addition, P. alcaliphila JAB1 is capable of cometabolically degrading cis-1,2-dichloroethylene (cDCE) when grown on phenol. The strain carries both catechol and protocatechuate branches of the β-ketoadipate pathway, which is used to funnel the pollutants to the central metabolism. Furthermore, we propose that clustering of MALDI-TOF MS spectra with closest phylogenetic relatives should be used when taxonomically classifying the isolated bacterium; this, together with 16S rRNA gene sequence and chemotaxonomic data analyses, enables more precise identification of the culture at the species level.
Electronic supplementary material
The online version of this article (10.1186/s40793-017-0306-7) contains supplementary material, which is available to authorized users.
Keywords: Pseudomonas alcaliphila JAB1; Pseudomonadaceae; Genome; Dioxygenase; Monooxygenase; Biodegradation; Bioremediation; Aromatic compounds; Biphenyl; Polychlorinated biphenyls (PCBs); Chlorobenzoic acids (CBAs); cis-1,2-dichloroethylene (cDCE); Phenol; bph genes; ben genes; phe genes; MALDI-TOF MS
Introduction
Over recent decades, significant quantities of potentially harmful chemicals have been released into the environment, creating countless numbers of contaminated sites. Major contaminants include halogenated and nitrated alicyclic, aliphatic, aromatic and polyaromatic compounds of industrial and agricultural origin. Many of these compounds have been found to have toxic, mutagenic and carcinogenic effects on living organisms. Removal of these xenobiotics usually involves physical and chemical processes, such as landfill, excavation and incineration, which are expensive and difficult to execute. An alternative approach, bioremediation, uses ubiquitous plant-microbe interactions to degrade xenobiotics [1]. Bacteria and fungi are natural recyclers capable of funneling toxic organic compounds to central metabolism intermediates [2], thereby creating harmless products [3] such as carbon dioxide and water. In addition to the enormous phylogenetic diversity of microorganisms, the richness of their metabolic activities promotes the degradation of pollutants and xenobiotics in different environments.
Members of the genus Pseudomonas [4, 5], one of the most diverse bacterial genera, inhabit several environmental niches and have been studied in relation to human and plant pathogenicity, antibiotic resistance, plant growth promotion, plant-derived organic matter degradation and bioremediation [6]. Pseudomonads, which are metabolically highly versatile, contain both abundant and unique metabolic pathways [7], which, most importantly, catabolize a broad range of substrates. Many of these substrates are pollutants, including aliphatic and aromatic petroleum hydrocarbons [8–12], BTEX [13, 14], phenolic compounds ranging from phenol via methylphenols and nitrophenols to chlorophenols [15], benzoate, CBAs and toluates [16], biphenyl and PCBs [17–19], chlorinated aliphatics [20] and many others. In this study, we present the first complete genome of the species P. alcaliphila, strain JAB1, whose extensive biodegradation capabilities are highlighted.
Organism information
Classification and features
P. alcaliphila was described as a facultatively psychrophilic alkaliphilic species isolated from seawater off the coast of Rumoi, Hokkaido, Japan. The characteristics of this species are as follows: alkaliphile, incapable of growth at >40 °C, catalase- and oxidase-positive and also capable of reducing nitrate to nitrite and of hydrolyzing casein and gelatin [21]. Further physiological features are listed in Table 1. The JAB1 cells are monotrichous rods as shown in Fig. 1.
Table 1.
Classification and general features of P. alcaliphila JAB1
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [55] | |
| Phylum Proteobacteria | TAS [56, 57] | ||
| Class Gammaproteobacteria | TAS [58, 59] | ||
| Order Pseudomonadales | TAS [5, 60] | ||
| Family Pseudomonadaceae | TAS [61] | ||
| Genus Pseudomonas | TAS [4, 5] | ||
| Species Pseudomonas alcaliphila | TAS [21] | ||
| Strain JAB1 (Accession no. DSM 26533) | |||
| Gram stain | Negative | TAS [21] | |
| Cell shape | Rod-shaped | IDA, TAS [21] | |
| Motility | Motile | IDA, TAS [21] | |
| Sporulation | Non-sporulating | TAS [21] | |
| Temperature range | Mesophile | IDA | |
| Optimum temperature | 28–37 °C | IDA | |
| pH range; Optimum | Not tested; Neutral | TAS [21] | |
| Carbon source | Biphenyl, phenol, other organic substrates | IDA | |
| MIGS-6 | Habitat | Soil | TAS [22] |
| MIGS-6.3 | Salinity | Up to 7% NaCl (w/v) | TAS [21] |
| MIGS-22 | Oxygen requirement | Aerobic | TAS [22] |
| MIGS-15 | Biotic relationship | Free-living | TAS [22] |
| MIGS-14 | Pathogenicity | Non-pathogen | NAS |
| MIGS-4 | Geographic location | Czech Republic | TAS [22] |
| MIGS-5 | Sample collection | 2000 | NAS |
| MIGS-4.1 | Latitude | 50°1′52″N | NAS |
| MIGS-4.2 | Longitude | 16°35′55″E | NAS |
| MIGS-4.4 | Altitude | 420 m | NAS |
IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence)
aEvidence codes
Fig. 1.
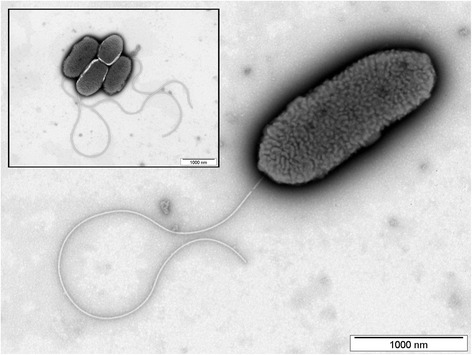
Transmission electron photomicrograph of P. alcaliphila JAB1
The JAB1 strain was originally misidentified as P. pseudoalcaligenes [22]. The consensus 16S rRNA gene sequence, compiled from four 16S rRNA gene copies contained in the JAB1 genome, had 99.93% similarity to those of P. alcaliphila AL 15-21T [21], P. chengduensis MBRT [23] and P. oleovorans subsp. lubricantis RS1T [24]. Additional closest matches included P. toyotomiensis HT-3T (99.86% similarity) and P. mendocina CH50T (99.24% similarity). A phylogenetic tree indicates closest relatedness of the JAB1 strain to P. alcaliphila AL 15-21T (Fig. 2). In addition, the JAB1 strain was unable to grow at 41 °C, which is a typical feature of P. alcaliphila but not of other closely related pseudomonads [23]. Furthermore, whole-cell MALDI-TOF MS analysis, performed following the methodology described elsewhere [12], indicated that JAB1 spectra clustered with those of P. alcaliphila AL 15-21T (Fig. 3). The results of MALDI-TOF MS profiling thus further confirmed the identity of the JAB1 strain as P. alcaliphila. Therefore, we propose that MALDI-TOF MS analysis be performed of the isolate and its closest phylogenetic relatives when taxonomically classifying the isolated bacterium. In addition to 16S rRNA gene sequence and chemotaxonomic data analysis, MALDI-TOF MS can provide additional information resulting in more precise classification of the isolate.
Fig. 2.
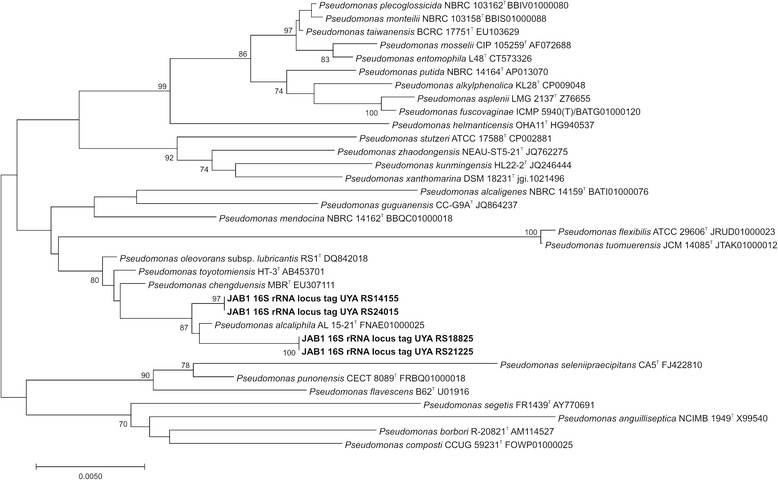
Phylogenetic tree indicating current phylogenetic placement of strain JAB1.The phylogenetic tree was constructed in MEGA7 [62] using secondary structure-based alignment of 16S rRNA gene sequences of the JAB1 strain (four copies of the 16S rRNA gene retrieved from the JAB1 genome) and its closest relatives [63]. The evolutionary distances were computed using the Kimura 2-parameter method [64]. All positions with less than 95% site coverage were eliminated; there were a total of 1375 positions in the final dataset. The tree construction method used was Neighbor-Joining [65]. The bootstrap test (1000 replicates) was used to test tree topology [66]; only bootstrap values >70 are indicated
Fig. 3.
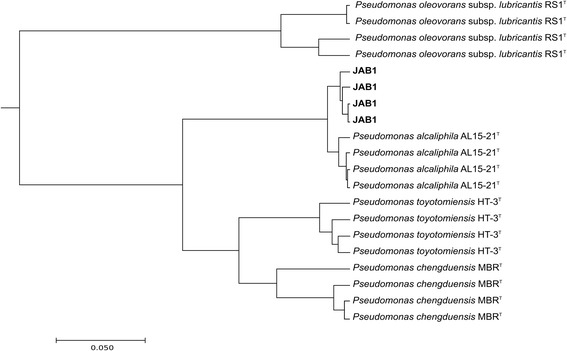
Clustering of MS spectra of strain JAB1 and related pseudomonads
Extended feature descriptions: biodegradative capabilities
The JAB1 strain was isolated from legacy contaminated soil in Jablonné nad Orlicí in the Czech Republic as a biphenyl-degrading bacterium capable of extensive degradation of several congeners of PCBs when grown in the presence of biphenyl [22]. PCB-degradation capabilities were further determined by a resting cell assay. Briefly, microcosms (volume of 119 ml) bearing biphenyl-preinduced P. alcaliphila JAB1 mid-log phase culture resuspended in mineral salt solution (20 ml per microcosm) were spiked with a 0.001% (w/v) commercial mixture of PCBs Delor 103 and incubated for 48 h. The content of individual PCB congeners present in the microcosms as well as CBA accumulation were determined using GC-MS (450-GC, 240-MS ion trap detector, Varian, Walnut Creek, CA). PCBs were analyzed in ethyl acetate extracts according to the method described by Čvančarová M. et al. [25]. CBAs were analyzed in the extracts using GC-MS after methylation with diazomethane according to our previously published protocol [26]. The respective chemical standards for the analytes were obtained from Merck (Darmstadt, Germany), Supelco (Steinheim, Germany), TCI Europe (Zwijndrecht, Belgium) and AccuStandard (New Haven, USA).
Metabolic activity of the JAB1 strain resulted in the depletion of various mono-, di-, tri- and tetra-chlorinated biphenyls as shown in Fig. 4. At the same time, 2-CBA, 3-CBA, 4-CBA, 2,3-diCBA, 2,4-diCBA and 2,5-diCBA formation (data not shown) was observed over the course of the coincubation period; these are common biodegradation intermediates of various PCB congeners and products of the upper PCB degradation pathway.
Fig. 4.
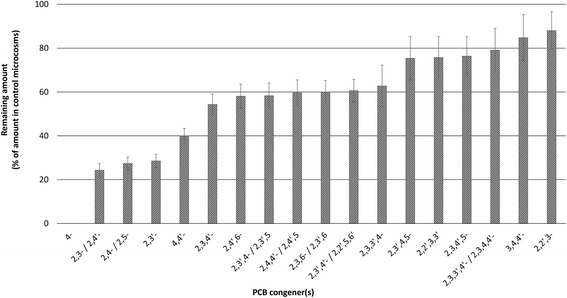
Degradation activities of strain JAB1 towards PCB congeners. Biphenyl-induced JAB1 cells were co-incubated with commercial PCB mixture Delor 103 for 48 h, with individual congener depletion being determined by GC-MS. Degradation of the following biphenyl derivatives was monitored but not observed: 2,2′-diCl, 2,2′,3-triCl, 2,2′,5-triCl, 2,2′,4-triCl, 2,2′,4,6′-tetraCl, 2,2′,3,6-tetraCl, 2,2′,3,6′-tetraCl, 2,2′,5,5′-tetraCl, 2,2′,4,5′-tetraCl, 2,2′,4,5 tetraCl, 2,2′,4,4′-tetraCl, 2,2′,3,5′-tetraCl, 2,2′,3,4′-tetraCl, 2,3′,4′,6-tetraCl, 2,2′,3,4 tetraCl, 2,3,4′,6-tetraCl, 2,4,4′,5-tetraCl, 2,3′,4′,5-tetraCl, 2,3′,4′,5′-tetraCl, 2,3′,4,4′-tetraCl, 3,3′,4,4′-tetraCl, 2,2′,3,4′,6-pentaCl, 2,2′,3,4,6′-pentaCl, 2,2′,4,5,5′-pentaCl, 2,3,3′,5′,6-pentaCl, 2,2′,4,4′,5 pentaCl, 2,2′,3,4′,5′-pentaCl, 2,2′,3,4,5′-pentaCl, 2,3,3′5,5′-pentaCl, 2,3,3′,4′,6-pentaCl, 2,2′,3,3′,4-pentaCl, 2,3′,4,4′,5-pentaCl, 2,3,3′,4,4′-pentaCl
The degradation of cDCE was assessed during growth on phenol, a compound known to promote cDCE degradation in microbial cultures [27]. Microcosms (volume of 119 ml) bearing P. alcaliphila JAB1 culture (20 ml per microcosm) grown and repeatedly propagated on 1 mM phenol and 0.01 mM cDCE were monitored for phenol and cDCE depletion as described elsewhere [28] as well as the increase in microbial culture density measured spectrophotometrically. Additional cultures were prepared in the presence of 0.01 mM cDCE as sole carbon source and of 0.01 mM cDCE and 1 mM sodium pyruvate to exclude direct utilization of cDCE by JAB1. In addition, SPME-GC-MS analysis of culture headspaces was performed over time in order to identify possible volatile chlorinated intermediates of the cDCE degradation pathway.
Monitoring of cDCE levels in cultures grown on 1 mM phenol revealed consistent cDCE depletion after the first 14 h of cultivation, when phenol (the growth-supporting substrate) was completely or almost completely consumed (Fig. 5). After 72 h of cultivation, 100% cDCE depletion was attained exclusively in phenol-grown cultures; during the same time period, cDCE loss accounted for only 13.4% ± 8.8 pp. in sodium pyruvate-grown cultures, 9.7% ± 9.1 pp. for JAB1 cultures exposed solely to cDCE and 12.5% ± 1.2 pp. in sterile medium. Such minimal cDCE loss under conditions tested can be ascribed to non-biological processes. Proven total cDCE depletion only in phenol-grown cultures confirms our hypothesis that cDCE degradation occurs cometabolically. To date, 1,1-dichloroacetaldehyde was confirmed as a degradation intermediate by means of SPM-GC-MS in culture headspaces. Quantification of this molecule and its accumulation over time will be an area for future research together with the study of possible metabolic pathways to promote further transformation of 1,1-dichloroacetaldehyde.
Fig. 5.
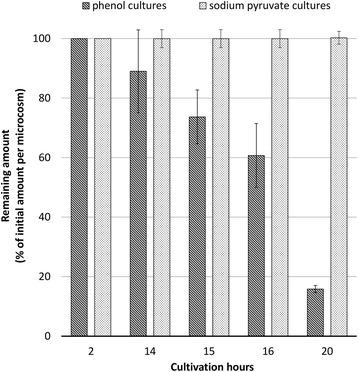
Trend in cDCE depletion over time in JAB1 cultures grown on 1 mM phenol or 1 mM sodium pyruvate in the presence of 0.01 mM cDCE. Residual amounts of cDCE over time are expressed as a percentage of initial cDCE content. Error bars indicate the standard deviation among three biological replicates
Genome sequencing information
Genome project history
The JAB1 strain was selected for sequencing due to its extensive degradative capabilities. The genome project was initiated in 2010. The complete genome sequence, deposited in GenBank under the accession number CP016162, was released public on April 14, 2017. A summary of the project information and its association with MIGS standard is shown in the Table 2.
Table 2.
Genome sequencing project information for P. alcaliphila JAB1
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | 454 shotgun, 454 8 kb paired-end |
| MIGS 29 | Sequencing platforms | GS FLX+ |
| MIGS 31.2 | Fold coverage | 41.5 |
| MIGS 30 | Assemblers | Newbler 2.8 |
| MIGS 32 | Gene calling method | GeneMarkS+ |
| Locus Tag | UYA | |
| Genbank ID | CP016162 | |
| GenBank Date of Release | April 14, 2017 | |
| GOLD ID | Gp0021677 | |
| BIOPROJECT | PRJNA104953 | |
| MIGS 13 | Source Material Identifier | DSM 26533 |
| Project relevance | Bioremediation, aromatic compounds degradation |
Growth conditions and genomic DNA preparation
Following its isolation from soil, the culture was preserved in a mineral salt solution [12] with biphenyl as sole carbon source and, over the long term, in glycerol stocks prepared from growing liquid cultures. For the purposes of DNA isolation, the culture was grown overnight on plate count agar (Difco, UK) at 28 °C. Genomic DNA was isolated using the PureLink™ Genomic DNA Mini Kit (Invitrogen, USA) according to the manufacturer’s instructions.
Genome sequencing and assembly
The genomic DNA of P. alcaliphila JAB1 was used to prepare shotgun and 8 kb paired-end 454 sequencing libraries according to the Library Preparation Method Manual (Roche). These libraries were sequenced with the GS FLX instrument using GS FLX+ chemistry (Roche) at the Institute of Molecular Genetics AS CR (Prague, Czech Republic). The resulting 110,702 shotgun and 461,976 paired-end reads comprised a total of 221,447,771 bases and represented 41.5-fold genome coverage. These reads were assembled in Newbler 2.8 software (Roche), and gaps were closed by local assembly of selected reads in Staden software [29] into a single circular contig.
Genome annotation
The NCBI Prokaryotic Genome Annotation Pipeline [30] was used for automated genome annotation, and functional annotations were carried out by searching against KEGG [31], COG [32] and Pfam [33] databases. SignalP [34] and TMHMM [35] tools were used for the prediction of genes with signal peptides and transmembrane helices, respectively. Searches against NCBI-NR [36], RefSeq [37], UniProtKB/Swiss-prot [38] and TCDB [39] were carried out for functional assignment of selected genes.
Genome properties
The P. alcaliphila JAB1 genome consists of a single 5,340,293 bp-long chromosome with a GC content of 62.5% (Fig. 6). The 4908 predicted genes correspond to an 89% coding density. The genome contains 4773 CDS, 65 tRNA genes for all 20 amino acids, 4 rRNA operons, 4 ncRNA genes, 3 CRISPR repeats and 54 pseudogenes. Function was assigned to 3816 CDS, with 957 CDS annotated as hypothetical proteins only (Table 3). The distribution of COG functional categories is shown in Table 4.
Fig. 6.
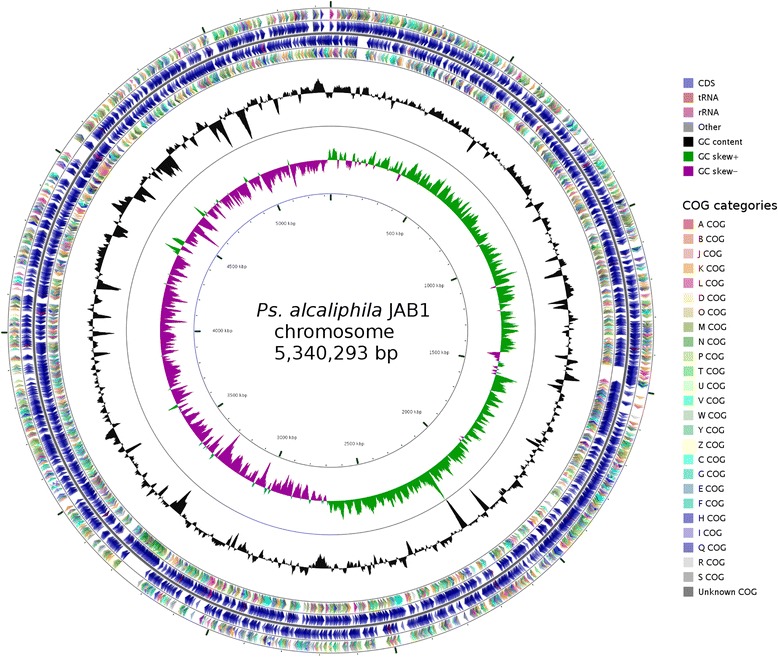
Circular map of the P. alcaliphila JAB1 chromosome. From outside to the center: CDS on forward strand colored according to their COG categories, all CDS and RNA genes on forward strand, all CDS and RNA genes on reverse strand, CDS on reverse strand colored according to their COG categories, GC content, and GC skew. The map was generated using CGView [67]
Table 3.
Genome statistics
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 5,340,293 | 100 |
| DNA coding (bp) | 4,749,316 | 88.93 |
| DNA G + C (bp) | 3,339,724 | 62.54 |
| DNA scaffolds | 1 | |
| Total genes | 4908 | 100 |
| Protein coding genes | 4773 | 97.25 |
| RNA genes | 81 | 1.65 |
| Pseudo genes | 54 | 1.10 |
| Genes in internal clusters | NA | |
| Genes with function prediction | 3816 | 77.75 |
| Genes assigned to COGs | 3927 | 80.01 |
| Genes with Pfam domains | 4221 | 86.00 |
| Genes with signal peptides | 557 | 11.35 |
| Genes with transmembrane helices | 1139 | 23.21 |
| CRISPR repeats | 3 | 0.06 |
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | %age | Description |
|---|---|---|---|
| J | 189 | 3.96 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.02 | RNA processing and modification |
| K | 368 | 7.71 | Transcription |
| L | 216 | 4.53 | Replication, recombination and repair |
| B | 2 | 0.04 | Chromatin structure and dynamics |
| D | 43 | 0.90 | Cell cycle control, Cell division, chromosome partitioning |
| V | 59 | 1.24 | Defense mechanisms |
| T | 425 | 8.90 | Signal transduction mechanisms |
| M | 232 | 4.86 | Cell wall/membrane biogenesis |
| N | 133 | 2.79 | Cell motility |
| U | 107 | 2.24 | Intracellular trafficking and secretion |
| O | 176 | 3.69 | Posttranslational modification, protein turnover, chaperones |
| C | 285 | 5.97 | Energy production and conversion |
| G | 196 | 4.11 | Carbohydrate transport and metabolism |
| E | 433 | 9.07 | Amino acid transport and metabolism |
| F | 88 | 1.84 | Nucleotide transport and metabolism |
| H | 179 | 3.75 | Coenzyme transport and metabolism |
| I | 189 | 3.96 | Lipid transport and metabolism |
| P | 256 | 5.36 | Inorganic ion transport and metabolism |
| Q | 118 | 2.47 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 574 | 12.03 | General function prediction only |
| S | 423 | 8.86 | Function unknown |
| – | 846 | 17.72 | Not in COGs |
The total is based on the total number of protein coding genes in the genome
Insights from the genome sequence
The genome harbors approximately 50 oxygenase genes, many of which are relevant to the biodegradation capabilities of the strain, especially those encoding for mono- and di-oxygenases responsible for the hydroxylation and opening of the aromatic ring, which are crucial steps in aromatic xenobiotic degradation (see below for details). Several other genes found are associated with the heavy metal resistance of the strain (e.g. heavy metal efflux P-type ATPases), indicating its overall adaptation to contaminated environments; organic compound contamination is often accompanied by high concentrations of heavy metals [40, 41]. The genome contains a complete set of genes for flagellar assembly and 50 additional genes associated with chemotaxis. Roughly 70 of the predicted CDS are putative intact or mutated transposase genes and phage-related genes.
Extended insights: degradation of aromatic compounds
In total, five regions harboring genes for aromatic compound degradation pathways were identified in the JAB1 genome as shown in Additional file 1: Table S1.
The first region, position range 1,474,745–1,486,552 in the annotated genome sequence, harbors genes encoding for a three-component benzoate 1,2-dioxygenase (benABC genes) followed by enzymes of a pathway enabling the complete degradation of benzoates and benzoate derivatives, yielding acetyl-CoA and pyruvate (catechol meta-cleavage pathway encoded by benD, fdx, and dmp cluster). The role played by the gene designated nahX encoding for putative ATP-cob(I)alamin adenosyltransferase in aromatic compound degradation is unclear [42]. No transcriptional regulator was found within or adjacent to this region, leading us to assume that genes from this region are not transcribed or their transcription is mediated in trans by regulatory proteins encoded elsewhere.
The second region, position range 1,518,873–1,540,279, harbors genes encoding for complete salicylate degradation via the catechol meta-cleavage pathway (sal/fdx genes), with salicylate 1-monooxygenase (encoded by salA) being responsible for opening the aromatic ring. Genes antAB encoding for large and small subunits of a putative terminal dioxygenase were also found in this region. Apart from genes presumed to be involved in the enzymatic degradation of aromatic compounds, ORFs encoding for transport proteins were found in this second region, namely a putative benzoate:H+ symporter (encoded by gene benE), a multi-component ABC transporter (six ORFs, designated ORF1–6) and a porin protein (benK), which are most likely involved in aromatic compound uptake. BenK ortholog has been shown to be involved in benzoate uptake in Acinetobacter sp. ADP1 [43]. Moreover, BenE and BenK orthologs from P. putida have been demonstrated to act as benzoate uptake proteins when heterologously expressed in yeasts [44]. Nevertheless, detailed experimental evidence is lacking on the function of these proteins and their role in aromatics/aromatic xenobiotics degradation, which represents a potential challenge for future research [45, 46].
The third region, position range 1,545,237–1,557,889, harbors bph genes encoding for enzymes of the complete upper and lower biphenyl degradation pathway. The structure of this operon is virtually identical to that of the bph operon found in the genome of the model PCB-degrading strain P. pseudoalcaligenes KF707 [47]. While pentadiene and its chlorinated derivatives can be degraded by enzymes encoded by the same operon (bphHIJ genes), no ORF encoding for benzoate terminal dioxygenase, essential for the completion of biphenyl degradation, is present in this region; nevertheless, such genes are found elsewhere in the JAB1 genome. Two other ORFs, tbuX and a gene designated ORF7, encoding for putative aromatic-transporter transmembrane proteins, were found in this region. Although the TbuX protein has been reported to be involved in the utilization of toluene in Ralstonia pickettii PKO1 [48], no detailed information exists on the precise role played by this group of proteins in aromatic compound transport. The predicted ORF7-encoded protein exhibits homology with members of the BphX family. Despite being commonly found in upper biphenyl degradation pathway-encoding supra-operonic clusters of various taxa, the function of these proteins in aromatic degradation is not known [49–51].
The fourth region, position range 4,397,470–4,411,693, harbors gene cluster pheKLMNOP encoding for multi-component phenol 2-monoxygenase and an fdx gene encoding a chloroplast-type ferredoxin essential for electron delivery to the active center of the oxygenase. The adjacent dmp cluster encodes for enzymes responsible for the meta-cleavage pathway of catechol/catechol derivatives, yielding acetyl-CoA and pyruvate. In this study, we demonstrate that strain JAB1 is capable of utilizing phenol as a sole carbon source and cometabolite of cDCE degradation.
The fifth region, position range 4,445,675 to 4,469,934, contains benABCD genes, which are responsible for the transformation of benzoate into catechol, as well as catABC and pcaDIJF genes, which encode the enzymes of the catechol branch of the β-ketoadipate pathway, yielding succinyl-CoA and acetyl-CoA [52]. This region also harbors pcaHGB genes for the protocatechuate branch of the β-ketoadipate pathway which enable hydroxybenzoate degradation; the pobA gene encoding 4-hydroxybenzoate 3-monooxygenase, responsible for the first step of 4-hydroxybenzoate hydroxylation, is located upstream of this fifth region (positions 4,435,851 to 4,437,041). In addition, this region harbors three ORFs encoding for putative transport proteins, which are most likely involved in aromatic compound uptake, designated as ORF8 (encoding for outer membrane porin from the OprD family), pcaK (an ortholog of p-hydroxybenzoate and protocatechuate transport protein from the Major Facilitator Superfamily [53]) and benE (benzoate:H+ symporter from the BenE family). Two other ORFs encoding for a cell division protein (zapE gene) [54] and a permease of unknown substrate specificity and function (designated ORF9) are also present in the fifth region in a pcaHG-zapE-ORF9 arrangement. As the intergenic region between the pcaG and zapE genes is only 1 bp in length, with zapE and ORF9 sequences even overlapping by 5 bp, we hypothesize that pcaHG-zapE-ORF9 represents a single transcriptional unit. The positioning of one or more genes involved in cell division as part of a cistron together with aromatic-degradation genes is rather unusual.
Since the regions harboring degradation-determining genes are flanked by transposase and conjugation protein-coding genes, and, at the same time, they generally exhibit analogous architecture and high similarity to corresponding orthologs from other pseudomonads, we hypothesize that their acquisition in the JAB1 genome was most likely due to multiple horizontal gene transfer events. However, further study is required to confirm this hypothesis.
Conclusions
Thorough microbiological experiments and whole genome sequence analysis lead us to conclude that P. alcaliphila JAB1 is a versatile degrader of organic pollutants. Its extensive degradative capabilities are enabled by a variety of genes determining the degradation of both biphenyl/biphenyl derivatives (mediated by the biphenyl degradation pathway encoded by the bph cluster) and monocyclic aromatics such as benzoate and its derivatives (halobenzoates, toluates), salicylic acid and phenol/phenolic compounds.
Acknowledgements
The authors wish to acknowledge Dr. Pavel Ulbrich for the transmission electron photomicrograph.
Funding
The research work involved in producing genomic and PCB degradation data was financed by grant no. 13-28283S, while the research into growth on phenol and cDCE degradation was funded by grant no. 14-32432S. Both grants were provided by the Czech Science Foundation, which played no role in designing the study, collecting, analyzing or interpreting the data or in writing the manuscript.
Abbreviations
- BTEX
Benzene, toluene, ethylbenzene, xylene
- CBAs
Chlorobenzoic acids
- cDCE
cis-1,2-dichloroethylene
- diCBA
Dichlorobenzoic acid
- GC-MS
Gas chromatography-mass spectrometry
- MALDI-TOF MS
Matrix-assisted laser ionization/desorption-time of flight mass spectrometry
- PCBs
Polychlorinated biphenyls
- pp
Percentage points
Additional file
Description of five regions harboring genes for aromatic compound degradation pathways were identified in the JAB1 genome. (DOCX 34 kb)
Authors’ contributions
Performed genome sequencing, assembly, annotation and analysis: JR, JS, MH and HS Performed microbiological experiments: JS, SF, MS, TC, AZ and OU Contributed reagents/materials: TM and HS Wrote the paper: JR, JS, SF and OU. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s40793-017-0306-7) contains supplementary material, which is available to authorized users.
Contributor Information
Hynek Strnad, Phone: (+420) 241063412, Email: strnad@img.cas.cz.
Ondrej Uhlik, Phone: (+420) 220445136, Email: ondrej.uhlik@vscht.cz.
References
- 1.Macková M, Dowling D, Macek T, editors. Phytoremediation and Rhizoremediation. Theoretical background. Dordrecht: Springer; 2006. [Google Scholar]
- 2.Eyers L, George I, Schuler L, Stenuit B, Agathos SN, El Fantroussi S. Environmental genomics: exploring the unmined richness of microbes to degrade xenobiotics. Appl Microbiol Biotechnol. 2004;66:123–130. doi: 10.1007/s00253-004-1703-6. [DOI] [PubMed] [Google Scholar]
- 3.Pieper DH, Seeger M. Bacterial metabolism of polychlorinated biphenyls. J Mol Microbiol Biotechnol. 2008;15:121–138. doi: 10.1159/000121325. [DOI] [PubMed] [Google Scholar]
- 4.Migula W. Arbeiten aus dem Bakteriologischen Institut der Technischen Hochschule zu Karlsruhe. Volume 1. 1895. Über ein neues System der Bakterien; pp. 235–238. [Google Scholar]
- 5.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Evol Microbiol. 1980;30:225–420. doi: 10.1099/00207713-30-1-225. [DOI] [Google Scholar]
- 6.Silby MW, Winstanley C, Godfrey SAC, Levy SB, Jackson RW. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev. 2011;35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- 7.Sørensen J, Nybroe O. Pseudomonas in the soil environment. In: Ramos J-L, editor. Pseudomonas: genomics, life style and molecular architecture. Boston: Springer US; 2004. pp. 369–401. [Google Scholar]
- 8.Kostka JE, Prakash O, Overholt WA, Green SJ, Freyer G, Canion A, Delgardio J, Norton N, Hazen TC, Huettel M. Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico Beach sands impacted by the Deepwater horizon oil spill. Appl Environ Microbiol. 2011;77:7962–7974. doi: 10.1128/AEM.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whyte LG, Bourbonniere L, Greer CW. Biodegradation of petroleum hydrocarbons by psychrotrophic Pseudomonas strains possessing both alkane (alk) and naphthalene (nah) catabolic pathways. Appl Environ Microbiol. 1997;63:3719–3723. doi: 10.1128/aem.63.9.3719-3723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn NW, Gunsalus IC. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol. 1973;114:974–979. doi: 10.1128/jb.114.3.974-979.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerniglia C. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation. 1992;3:351–368. doi: 10.1007/BF00129093. [DOI] [Google Scholar]
- 12.Wald J, Hroudová M, Jansa J, Vrchotová B, Macek T, Uhlík O. Pseudomonads rule degradation of polyaromatic hydrocarbons in aerated sediment. Front Microbiol. 2015;6:1268. doi: 10.3389/fmicb.2015.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Applegate BM, Kehrmeyer SR, Sayler GS. A chromosomally based tod-luxCDABE whole-cell reporter for benzene, toluene, ethybenzene, and xylene (BTEX) sensing. Appl Environ Microbiol. 1998;64:2730–2735. doi: 10.1128/aem.64.7.2730-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baldwin BR, Mesarch MB, Nies L. Broad substrate specificity of naphthalene- and biphenyl-utilizing bacteria. Appl Microbiol Biotechnol. 2000;53:748–753. doi: 10.1007/s002539900300. [DOI] [PubMed] [Google Scholar]
- 15.Spain JC, Gibson DT. Oxidation of substituted phenols by Pseudomonas putida F1 and Pseudomonas sp. strain JS6. Appl Environ Microbiol. 1988;54:1399–1404. doi: 10.1128/aem.54.6.1399-1404.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez BS, Higson FK, Kondrat R, Focht DD. Metabolism of and inhibition by chlorobenzoates in Pseudomonas putida P111. Appl Environ Microbiol. 1991;57:3361–3366. doi: 10.1128/aem.57.11.3361-3366.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furukawa K, Miyazaki T. Cloning of a gene cluster encoding biphenyl and chlorobiphenyl degradation in Pseudomonas pseudoalcaligenes. J Bacteriol. 1986;166:392–398. doi: 10.1128/jb.166.2.392-398.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nováková H, Vošáhlíková M, Pazlarová J, Macková M, Burkhard J, Demnerová K. PCB metabolism by Pseudomonas sp. P2. Int Biodeterior Biodegr. 2002;50:47–54. doi: 10.1016/S0964-8305(02)00058-6. [DOI] [Google Scholar]
- 19.Leigh MB, Prouzová P, Macková M, Macek T, Nagle DP, Fletcher JS. Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl Environ Microbiol. 2006;72:2331–2342. doi: 10.1128/AEM.72.4.2331-2342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wackett LP, Gibson DT. Degradation of trichloroethylene by toluene dioxygenase in whole-cell studies with Pseudomonas putida F1. Appl Environ Microbiol. 1988;54:1703–1708. doi: 10.1128/aem.54.7.1703-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yumoto I, Yamazaki K, Hishinuma M, Nodasaka Y, Suemori A, Nakajima K, Inoue N, Kawasaki K. Pseudomonas alcaliphila sp. nov., a novel facultatively psychrophilic alkaliphile isolated from seawater. Int J Syst Evol Microbiol. 2001;51:349–355. doi: 10.1099/00207713-51-2-349. [DOI] [PubMed] [Google Scholar]
- 22.Ryšlavá E, Krejčík Z, Macek T, Nováková H, Demnerová K, Macková M. Study of PCB degradation in real contaminated soil. Fresenius Environ Bull. 2003;12:296–301. [Google Scholar]
- 23.Tao Y, Zhou Y, He X, Hu X, Li D. Pseudomonas chengduensis sp. nov., isolated from landfill leachate. Int J Syst Evol Microbiol. 2014;64:95–100. doi: 10.1099/ijs.0.050294-0. [DOI] [PubMed] [Google Scholar]
- 24.Saha R, Spröer C, Beck B, Bagley S. Pseudomonas oleovorans subsp. lubricantis subsp. nov., and reclassification of Pseudomonas pseudoalcaligenes ATCC 17440T as later synonym of Pseudomonas oleovorans ATCC 8062T. Curr Microbiol. 2010;60:294–300. doi: 10.1007/s00284-009-9540-6. [DOI] [PubMed] [Google Scholar]
- 25.Čvančarová M, Křesinová Z, Filipová A, Covino S, Cajthaml T. Biodegradation of PCBs by ligninolytic fungi and characterization of the degradation products. Chemosphere. 2012;88:1317–1323. doi: 10.1016/j.chemosphere.2012.03.107. [DOI] [PubMed] [Google Scholar]
- 26.Křesinová Z, Hostačná L, Medková J, Čvančarová M, Stella T, Cajthaml T, Sensitive GC. MS determination of 15 isomers of chlorobenzoic acids in accelerated solvent extracts of soils historically contaminated with PCBs and validation of the entire method. Int J Environ Anal Chem. 2014;94:822–836. doi: 10.1080/03067319.2014.900677. [DOI] [Google Scholar]
- 27.Hopkins GD, McCarty PL. Field evaluation of in situ aerobic cometabolism of trichloroethylene and three dichloroethylene isomers using phenol and toluene as the primary substrates. Environmen Sci Technol. 1995;29:1628–1637. doi: 10.1021/es00006a029. [DOI] [PubMed] [Google Scholar]
- 28.Fraraccio S, Strejček M, Dolinová I, Macek T, Uhlík O. Secondary compound hypothesis revisited: selected plant secondary metabolites promote bacterial degradation of cis-1,2-dichloroethylene (cDCE) Sci Rep. 2017;7:8406. doi: 10.1038/s41598-017-07760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staden R. The staden sequence analysis package. Mol Biotechnol. 1996;5:233. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 30.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN. The COG database: an updated version includes eukaryotes. BMC bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 35.Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 36.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2010;38:D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2015;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saier MH, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. The transporter classification database (TCDB): recent advances. Nucleic Acids Res. 2016;44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roane T, Josephson K, Pepper I. Dual-bioaugmentation strategy to enhance remediation of cocontaminated soil. Appl Environ Microbiol. 2001;67:3208–3215. doi: 10.1128/AEM.67.7.3208-3215.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gremion F, Chatzinotas A, Kaufmann K, Von Sigler W, Harms H. Impacts of heavy metal contamination and phytoremediation on a microbial community during a twelve-month microcosm experiment. FEMS Microbiol Ecol. 2004;48:273–283. doi: 10.1016/j.femsec.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Sota M, Yano H, Ono A, Miyazaki R, Ishii H, Genka H, Top EM, Tsuda M. Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its Transposon Tn4655 suggests catabolic gene spread by a tyrosine Recombinase. J Bacteriol. 2006;188:4057–4067. doi: 10.1128/JB.00185-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collier LS, Nichols NN, Neidle EL. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J Bacteriol. 1997;179:5943–5946. doi: 10.1128/jb.179.18.5943-5946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishikawa Y, Yasumi Y, Noguchi S, Sakamoto H, Nikawa J. Functional analyses of Pseudomonas putida benzoate transporters expressed in the yeast Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 2008;72:2034–2038. doi: 10.1271/bbb.80156. [DOI] [PubMed] [Google Scholar]
- 45.Cowles CE, Nichols NN, Harwood CS. BenR, a XylS homologue, regulates three different pathways of aromatic acid degradation in Pseudomonas putida. J Bacteriol. 2000;182:6339–6346. doi: 10.1128/JB.182.22.6339-6346.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang DS, Whang TJ, Cheng FC, YP W, Wang YT, Luo WI, Wang YS. Toxicity assessment of mono-substituted benzenes and phenols using a Pseudomonas initial oxygen uptake assay. Environ Toxicol Chem. 2005;24:253–260. doi: 10.1897/04-212R.1. [DOI] [PubMed] [Google Scholar]
- 47.Taira K, Hirose J, Hayashida S, Furukawa K. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J Biol Chem. 1992;267:4844–4853. [PubMed] [Google Scholar]
- 48.Kahng H-Y, Byrne AM, Olsen RH, Kukor JJ. Characterization and role of tbuX in utilization of toluene by Ralstonia pickettii PKO1. J Bacteriol. 2000;182:1232–1242. doi: 10.1128/JB.182.5.1232-1242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li A, YY Q, Pi WQ, Zhou JT, Gai ZH, Xu P. Metabolic characterization and genes for the conversion of biphenyl in Dyella ginsengisoli LA-4. Biotechnol Bioeng. 2012;109:609–613. doi: 10.1002/bit.23333. [DOI] [PubMed] [Google Scholar]
- 50.Furukawa K, Suenaga H, Goto M. Biphenyl dioxygenases: functional versatilities and directed evolution. J Bacteriol. 2004;186:5189–5196. doi: 10.1128/JB.186.16.5189-5196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romine MF, Stillwell LC, Wong K-K, Thurston SJ, Sisk EC, Sensen C, Gaasterland T, Fredrickson JK, Saffer JD. Complete sequence of a 184-Kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J Bacteriol. 1999;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harwood CS, Parales RE. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 53.Nichols NN, Harwood CS. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J Bacteriol. 1997;179:5056–5061. doi: 10.1128/jb.179.16.5056-5061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marteyn BS, Karimova G, Fenton AK, Gazi AD, West N, Touqui L, Prevost M-C, Betton J-M, Poyraz O, Ladant D, et al. ZapE is a novel cell division protein interacting with FtsZ and modulating the Z-ring dynamics. MBio. 2014;5:e00022–14. [DOI] [PMC free article] [PubMed]
- 55.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, bacteria, and Eucarya. Proc Natl Acad Sci. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stackebrandt E, Murray RGE, Trüper HG. Proteobacteria classis nov., a name for the Phylogenetic Taxon that includes the “purple bacteria and their relatives”. Int J Syst Evol Microbiol. 1988;38:321–325. [Google Scholar]
- 57.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. Nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, Boone DR, De Vos P, Goodfellow M, Rainey FA, Schleifer K-H, editors. Bergey’s manual of systematic bacteriology: volume two the Proteobacteria, part B the Gammaproteobacteria. Boston: Springer; 2005. pp. 323–442. [Google Scholar]
- 58.Euzéby J. Validation of publication of new names and new combinations previously effectively published outside the IJSEM. Int J Syst Evol Microbiol. 2005;55:2235–2238. doi: 10.1099/ijs.0.64108-0. [DOI] [PubMed] [Google Scholar]
- 59.Garrity GM, Bell JA, Lilburn T. Class III. Gammaproteobacteria class. Nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, Boone DR, De Vos P, Goodfellow M, Rainey FA, Schleifer K-H, editors. Bergey’s manual of systematic bacteriology: volume two the Proteobacteria, part B the Gammaproteobacteria. Boston: Springer; 2005. pp. 323–442. [Google Scholar]
- 60.Orla-Jensen S. The main lines of the natural bacterial system. J Bacteriol. 1921;6:263–273. doi: 10.1128/jb.6.3.263-273.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garrity GM, Bell JA, Lilburn T. Pseudomonadaceae Winslow, Broadhurst, Buchanan, Krumwiede, Rogers and Smith 1917, 555AL. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, Boone DR, De Vos P, Goodfellow M, Rainey FA, Schleifer K-H, editors. Bergey’s manual of systematic bacteriology: volume two the Proteobacteria, part B the Gammaproteobacteria. Boston: Springer; 2005. pp. 323–411. [Google Scholar]
- 62.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim M, HS O, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 64.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 65.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 66.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 67.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]


