Fig. 4.
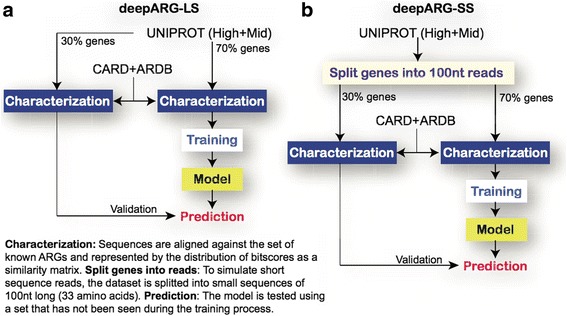
Classification framework. UNIPROT genes were used for validation and training whereas the CARD and ARDB databases were used as features. The distance between genes from UNIPROT to ARGs databases is computed using the sequence alignment bit score. Alignments are done using DIAMOND with permissive cutoffs allowing a high number of hits for each UNIPROT gene. This distribution is used to train and validate the deep learning models (The panel in the figure provides additional description on the training of the models)
