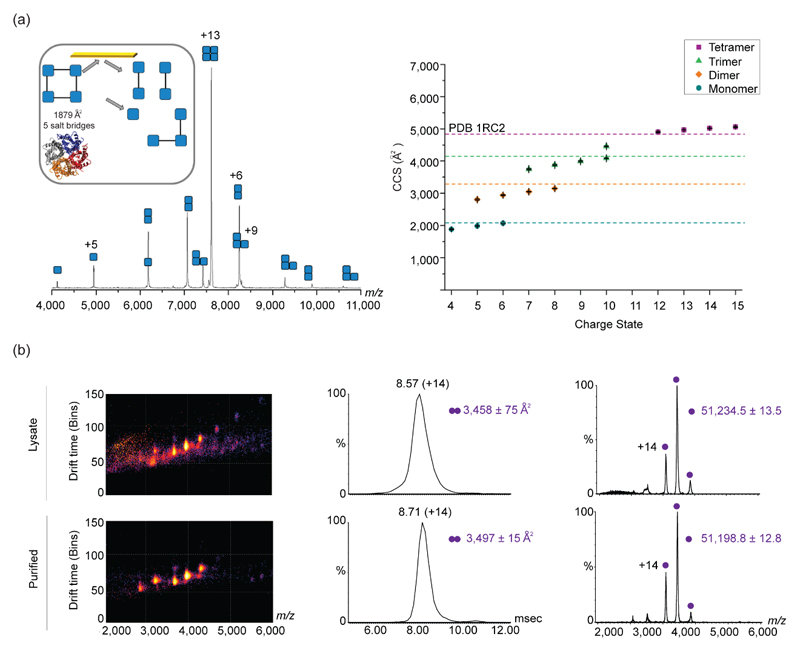
Figure 2. Applying IM-MS for structural characterization of soluble and membrane proteins.
(a) Application of SID with IM-MS for the study of membrane proteins is shown for AqpZ. The SID spectrum for the 13+ charge state of AqpZ, shows the different subcomplexes that were dissociated after collision with the surface. The inset represents the structure of AqpZ and the interfacial analysis, as determined from the PISA software. The crystal structure (PDB 1RC2) is shown below. The right graph shows the CCS distribution of the different sub-complexes of AqpZ. Theoretical CCS values of the different structures, represented as dashed lines, were determined using MOBCAL and the scaled PA. Adapted with permission from [21]•. (b) IM-MS analysis of recombinant proteins directly from crude cell lysates produces comparable information to that obtained from purified proteins, as exemplified for Hsp31. The three-dimensional IM-MS spectra of the crude cell lysate and purified samples are shown on the left, while the arrival time distribution and two dimensional spectra of m/z over intensity are shown on the middle and right panels, respectively. Adapted with permission from [22]•.