Extended Data Figure 4. NL CAMs control neuron-induced astrocyte morphogenesis.
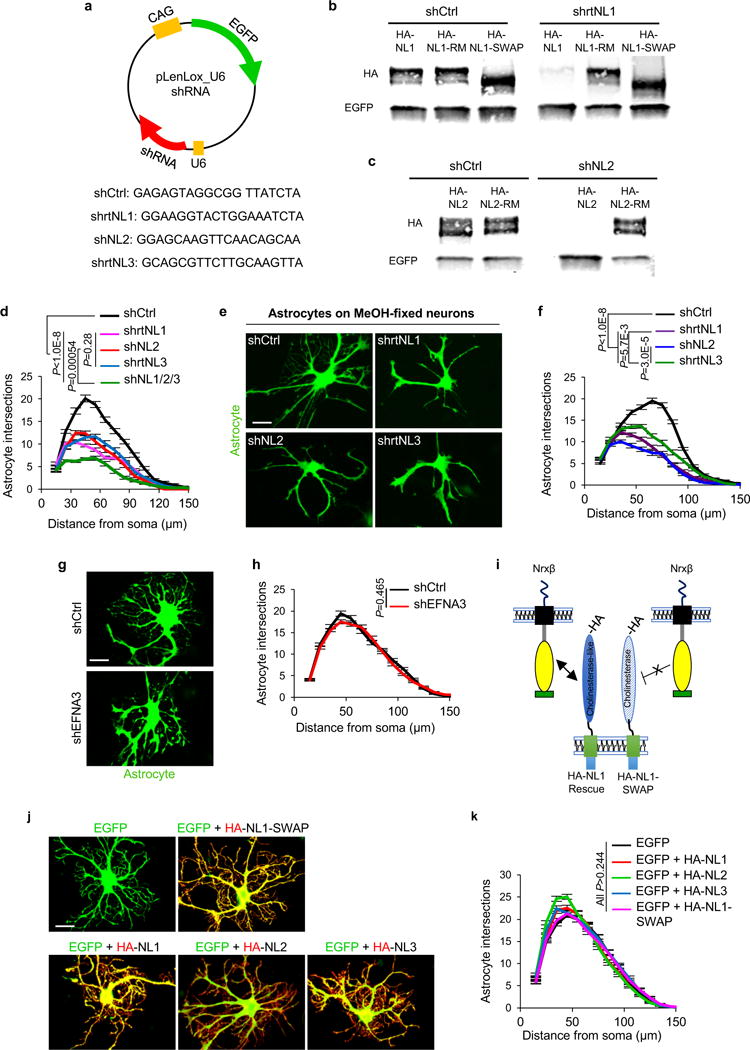
a-c, Specificity of shRNAs against NL sequences. a, Schematic representation of EGFP-expressing shRNA plasmids used to silence rat NLs in vitro. Sequences of shRNAs used on rat astrocytes against NL1, NL2, and NL3, as well as a scrambled shNL1 sequence used as a control (shCtrl). Rat-specific shRNAs against NL1, NL2, and NL3 were previously verified for efficiency and specificity by42,43 shNL2 sequence matches both rat and mouse NL2 sequences and effectively silences NL2 expression in cells from both species (see Extended Data Fig. 6c-d). b, Western blot analyses of cell lysates from HEK293 cells transfected with shCtrl or shrtNL1 with various HA-tagged NL1 constructs. Full length NL1 (HA-NL1) is effectively silenced by the shrtNL1, whereas NL1 shRNA-resistant mutant (HA-NL1-RM) and NL1-SWAP are not silenced by shrtNL1. Note: NL1-SWAP runs smaller than full-length NL1 (previously published in23). c, Western blot analyses of cell lysates from HEK293 cells transfected with shCtrl or shNL2 with HA-tagged NL2 or HA-tagged NL2-RM. Full length NL2 (HA-NL2) is effectively silenced by the shNL2, whereas NL2-RM is not. b, c, Antibodies against HA were used to detect the expression of NLs and the lysates were blotted for using an EGFP-specific antibody to verify transfection with shRNA plasmids. d-k, NLs play significant roles in controlling neuron-induced astrocyte morphogenesis. d, Sholl quantification of astrocyte complexity of NL1, NL2, NL3, or NL1/2/3 silenced astrocytes cultured on cortical neurons (not visible, compare with Fig. 2a-e). Data represent 1 experiment with 3 biological replicates. Similar results were obtained in 3 independent experiments. n>25cells/condition/experiment. e, Representative images of shNL-transfected astrocytes (green) cultured with MeOH-fixed neurons (not visible). f, Sholl quantification of astrocyte complexity of NL-silenced astrocytes cultured on MeOH-fixed neurons. Data represent 1 experiment with 3 biological replicates. Similar results were obtained in 2 independent experiments. n>20 cells/condition/experiment. g, Representative images of transfected astrocytes (green) with shCtrl or an shRNA against EphrinA3 (EFNA3) in co-culture with neurons (not visible). EFNA3 is a CAM expressed in astrocytes that regulates astrocyte-synapse interactions in the hippocampus22. h, Sholl quantification of astrocyte complexity in shCtrl and shEFNA3-transfected astrocytes in co-culture with neurons. Data represent 1 experiment with 3 biological replicates. Similar results were obtained in 2 independent experiments. n>20cells/condition/experiment. i, Cartoon representation of NL1 domain structure. NLs are type-I transmembrane proteins with a large N-terminal extracellular domain (ECD). NL1-ECD contains a cholinesterase (ChoE)-like domain that interacts with Nrxβs transcellularly. In a previous study, a chimera of NL1 (NL1-SWAP) was created in which the ChoE-like domain is swapped for the ChoE sequence. This chimera is efficiently trafficked to the cell surface, but fails to interact transcellularly with Nrx-βs23. j, Representative images of astrocytes over-expressing EGFP (green) and HA-tagged NLs (red) in co-culture with neurons (not visible). NL over-expressing astrocytes do not show any reductions in neuron-induced astrocyte morphogenesis. k, Sholl quantification of astrocyte complexity when over-expressing HA tagged-NLs. Data represent 1 experiment with 3 biological replicates. Similar results were obtained in 2 independent experiments. n>20 cells/condition/experiment. ANCOVA (d, f, h, k). For gel source data, see Supplementary Figure 1. Data are means ± s.e.m. Scale bars, 10 μm.
