Abstract
Bortezomib (BTZ) is highly effective in the treatment of Multiple Myeloma (MM), however emergent drug resistance is common. Consequently, we employed CRISPR targeting 19,052 human genes to identify unbiased targets that contribute to BTZ resistance. Specifically, we engineered an RPMI8226 MM cell line to express Cas9 and a lentiviral-vector CRISPR library and cultured derived cells in doses of BTZ lethal to parental cells. Sequencing was performed on surviving cells to identify inactivated genes responsible for drug resistance. From two independent whole genome screens, we selected 31 candidate genes and constructed a second CRISPR sgRNA library, specifically targeting each of these 31 genes with four sgRNAs. After secondary screening for BTZ resistance, the top 20 “resistance” genes were selected for individual validation. Of these 20 targets the proteasome regulatory subunit PSMC6 was the only gene validated to reproducibly confer BTZ resistance. We confirmed that inhibition of chymotrypsin-like proteasome activity by BTZ was significantly reduced in cells lacking PSMC6. We individually investigated other members of the PSMC group (PSMC1 to 5) and found that deficiency in each of those subunits also imparts BTZ resistance. We found 36 mutations in 19S proteasome subunits out of 895 patients in the IA10 release of the CoMMapss study (https://www.themmfr.org/). Our findings demonstrate that the PSMC6 subunit is the most prominent target required for BTZ sensitivity in MM cells and should be examined in drug refractory populations.
Keywords: Proteasome, PSMC6, multiple myeloma, Bortezomib resistance
Introduction
Multiple myeloma (MM) is a plasma cell malignancy, which has seen significant improvements in outcomes following the introduction of proteasome inhibitors (PI), specifically bortezomib (BTZ), carfilzomib and ixazomib into routine clinical practice (1–6). PIs used in combination with other compounds, achieve initial response in nearly 90–100% of patients (7). Despite the high initial response rates, almost all patients develop drug resistance over time and eventually becoming refractory to PIs. The underlying mechanisms of such PI resistance are still incompletely understood (8). The genes and pathways most commonly associated with BTZ resistance are PSMB5 (9–13), the ERN1-XBP1 pathway (14, 15) as well as the IGF-1 axis (16). PSBM5 mutations have been identified frequently in MM cell line but are not found in newly diagnosed patients (17, 18) while XBP-1 mutations have also been seen but are also found in less than 1% of newly diagnosed MM cases. Indeed the mutation prevalence of these genes combined is only 2% in supposedly PI refractory patients (19). XBP1 knock down experiments have shown correlation with BTZ resistance (14, 20). Recently, Acosta-Alvear et al reported an shRNA screening and found that genetic depletion of the vast majority of the 19S proteasomal regulator subunits including PSMC1 and 6, conferred a marked resistance to PIs (21). In a separate study Tsvetkov et al using gene trap screening identified that compromising the 19S proteasome including PSMC2 - 6 protected cells from proteasome inhibitors.(22) Sheffer et al.(23) using CRISPR technology also identified loss of PSMC6 as inducing resistance although to date results are only published in abstract form.
CRISPR-Cas9 knockout is a precise and efficient genome-editing technology (24, 25). This simple gene-engineering tool consists of a 20-base pair single-guide RNA (sgRNA) and a Cas9 protein, which can specifically recognize and cut the sgRNA binding region (24). Recently, a genome-scale CRISPR-Cas9 library was developed (26, 27) including 19,052 human genes, each gene being targeted with six sgRNAs. We used this genome-wide library to identify genes that are potentially associated with BTZ resistance. Our screening identified absence of the proteasome regulatory subunit PSMC6 as gene most consistently conferring BTZ resistance in human multiple myeloma cells.
Materials and Methods
Cell lines and plasmids
Human multiple myeloma cell lines (HMCLs) RPMI8226, FR4, JJN3, KMS11, KMS18, MM1.R, OPM2, and U266 were maintained in RPMI1640 with 5% fetal bovine serum, 100u/ml penicillin and 100 ug/ml streptomycin. All MM cells were from Dr. Leif Bergsagel’s lab. These cells were tested for mycoplasma negative and validated by finger printing develop by Dr. Bergsagel’s lab (unpulished data). All experiments were performed on MM cells less than 20 passages after thawed from liquid nitrogen. Plasmids pCDHpuroCr expressing selection marker puromycin and pCDHgfpCr expressing a green fluorescent protein selection marker (GFP) for cloning sgRNAs were derived from pCDH-CMV-MCS-EF1-Puro and pCDH-CMV-MCS-EF1-copGFP (SBI, Mountain View, CA 94043) respectively by cloning the sgRNA expression cassette from plasmid lentiGuide-Puro (Addgene #52963).(28). Plasmid pCDHBlastCas9HA was derived from lentiCas9-blast (#52962).(28) by cloning Cas9 gene (Flag tag was replaced with HA tag) into pCDHBlast under the control of CMV promoter, which was constructed by Shi et al. (C.X. Shi; unpublished observations).
Small Molecules
Tunicamycin was from Abcam (Cat. Ab120296, Cambridge, MA, USA 02139) and its structure was published by Takatsuki et al(29). Staurosporine was from ApexBio (Cat. A8192, Houston TX USA 77054) and its structure was published by Furusaki et al (30, 31). BTZ was from LC laboratories (Cat. B-1408, Wobum, MA USA 01801). Carfilzomib was from MCE Medchem Express (Cat. HY10455, Monmouth Junction, NJ USA 08852). Dexamethasone (Cat. D4902-25MG) and melphalan (Cat M2011-100MG) were from Sigma (St. Louis, MO USA, 63103).
Utilization of sgRNA
The GeCKO V2 library (Addgene #100000049)(28) was expanded by transforming the library into DH10B cells by electrophoresis with a total of 2.5X107 clones produced. A sgRNA library for the second round screening was created by cloning sgRNAs synthesized by IDT technology (Coralville, Iowa USA 52241) individually, mixed manually and finally cloned into the vector pCDHgfpCr. All other individual sgRNAs were cloned either into the plasmid pCDHpuroCr or pCDHgfpCr. sgRNA sequences used for targeting PSMC1 to 6 are displayed in supplemental 1.
Lenti-vector packaging and infection
A modified Graham’s calcium-phosphate method (32, 33) was used for packaging lentiviral vectors. Briefly, 293T cells were split into 100 mm tissue culture dishes one to four (total volume 15 ml medium, no more than 70% confluent before transformation). Transformation was performed 24 hours later. For construction, 2ml calcium phosphate-DNA co-precipitation solution was added to two packaging plasmids (18.34 ug for pSPAX2, 7.34 ug for pMD2.G) and a lenti vector (24.34 ug), 100 ul 2.5M CaCl2, add 1/10 TE, PH8.0 to total volume to 1 ml. Then one volume of 2xCa/DNA solution was added quickly to an equal volume of 2x HEPES solution. After 1 minute, 1.5 ml of the mixed solution was added to the cell culture medium. The next morning the medium was changed with 12 ml fresh DMEM medium with 10% FBS. The viruses were harvested 20 hours after medium change. For infection of MM cells one million cells were added into six well plates with 6 ul polybrene (5 mg/ml) and 1.5 ml lenti virus and added fresh medium to total volume 3ml. One hour later, 3 ml of fresh medium was added. Next morning the medium was changed to 6 ml fresh medium.
CRISPR sgRNA screening
RPMI8226Cas9 cells were infected with the CRISPR library. The infected cells were treated with BTZ at 7nM for seven days (changed medium with 7 nM BTZ at day 2 and day 4). Residual cells surviving BTZ treatment were expanded over 15 days without BTZ treatment and then subjected to DNA sequencing
DNA sequencing and data analysis
We screened the abundance of sgRNA targeting the ~19K genes in cells showing resistance to BTZ by next generation sequencing (Ion Torrent PGM, ThermoFisher). An in house algorithm was used to clean the gene-specific sgRNA by eliminating the surrounding adapters sequences. A script written in the Perl programming language was used to identify all instances of the “tag” sequence AAAGGACGAAACACC? that occur in the fasta files. The ‘?’ can denote A, C, G, or T. The script also captures the next “target” sequence of twenty nucleotides downstream from the tag. The targets are stored in a hash table which counts the frequency with which each target appears adjacent to the tag. The gene-specific sgRNA were quantified and the 100 most abundant hits were subsequently blasted against the human genome.
Proteasome chymotrypsin-like activity, caspase 3&7 activity assay and 26S proteasome assay
Proteasome chymotrypsin-like activity was measured by cell based assay which was performed according manufacture description (Cat No. G8660, Promega. Madison, WI, USA 53711). Briefly, 2,000 cells were plated into 384-well plates. Cells were treated with different concentration (3, 4, 5, 6, 7, 8 and 9 nM) of BTZ for 24 hours. Then, proteasome-Glo reagent was added into the cells. Luciferase activity was assayed by luciferase reader Cytation 3 imaging reader (Winooski, VT 05404 USA). Caspase 3 & 7 activity assay was similar to proteasome chymotrypsin-like activity but using caspase 3 & 7 Glo (Cat No. G8091 Madison, WI, USA 53711) instead of proteasome-Glo. Total 26S proteasome assay was performed by ELISA kit according the manufacture instruction (Cat No. 353541, United States Biological, Salem, MA USA 01970). This kit is specific only for the detection of the intact 26S proteasome in cell lysates. The capture antibody is specific for the 20S particle while the detection antibody is specific for the 19S particle. This kit is not cross-reactive with the following: 19S particle, 20S particle, PSMB1, LMP2 and PSMA3.
Western Blot and MTT assay
We used standard Western Blot. HA antibody (Cat. No. MMS-101P Clone 16B12) was from Covance (Emeryville California USA 94608). Caspase 8 (Cat. 9746S), ERN1 (Cat. 9956), SQSTM1/p62 (D5E2)(Cat. 8025S) and VCP/p97 (Cat. 2648) antibodies were from Cell Signaling (Danvers MA USA 01923). UFD1 antibody was from Bethyl Laboratories Inc (Montgomery, TX Cat. A303-847A). Immunoblot quantification was performed by the software ImageJ (National Institutes of Health, Bethesda, MD.(34) MM cells viability was determined by MTT assay (Sigma-Aldrich) following the manufacturer’s instructions.
Quantitative PCR
Total RNA was isolated using RNeasy Plus Mini kit (QIAGEN) and reverse-transcribed using QuantiTect Reverse Transcription kit (QIAGEN). Quantitative PCR was performed using TaqMan Universal PCR Master Mix with predesigned probe (Applied Biosystems), and the comparative CT method was used for relative quantification on an ABI 7900HT Fast Real-Time PCR system (Applied Biosystems).
Results
Targeting MM cells with CRISPR-sgRNA
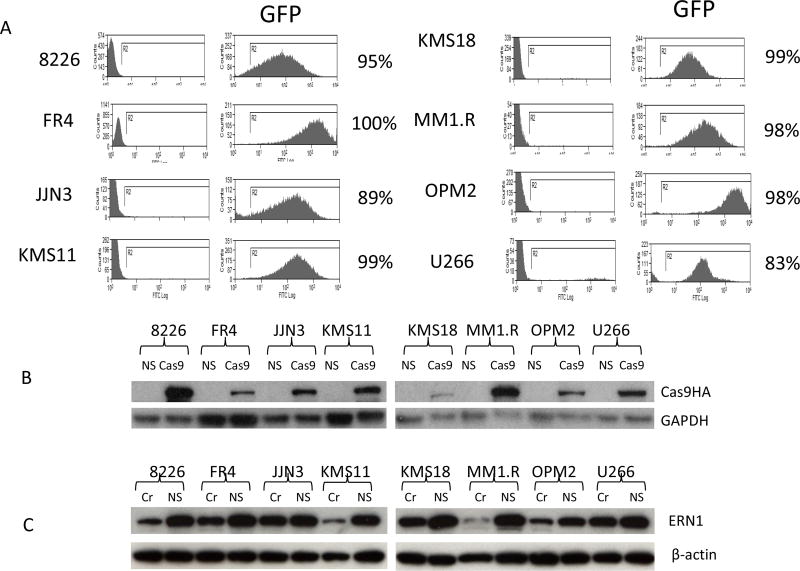
We first tested infection efficiency in eight MM cell lines (RPMI8226, KMS11, KMS18, FR4, JJN3, MM1.R, OPM2 and U266) with a lentivirus bearing a GFP marker (packaged from pCDH-CMV-MCS-EF1-copGFP). Flow cytometry results indicated that all cell lines were sensitive to lentivirus infection (Figure 1A). We then engineered all eight cell lines to over express Cas9 with a HA tag using lentivectors packaged from plasmid pCDHBlastCas9HA under selection of blasticidin for two weeks. We analyzed Cas9 expression of these eight cell lines. RPMI8226, MM1.R and KMS11 expressed higher levels of Cas9 than the remaining five cell lines (Figure 1B). Next, we tested knockout efficiency using a sgRNA targeting ERN1 in all eight cell lines. ERN1 expression level was significantly reduced in the three cell lines showing higher Cas9 level (MM1.R, KMS11, and RPMI8226) (Figure 1C). RPMI8226 cells were selected for primary sgRNA whole genome library screening, because of their superior capability for colony formation when compared to MM1.R and KMS11.
Figure 1.
Infecetion efficiency of lentivirus and targeting efficiency of sgRNA against ERN1 in MM cells. (A) Lenti virus infection efficiency to MM cells detected by flow cytometry. (B). Cas9 with HA tag was detected by Western Blot with HA antibody. (C) ERN1 knocking out efficiency by CRISPR technology in MM cells. Cr=ERN1 sgRNA. NS= non-specific sgRNA control.
PSMC6 was identified as the only reproducible gene conferring BTZ resistance
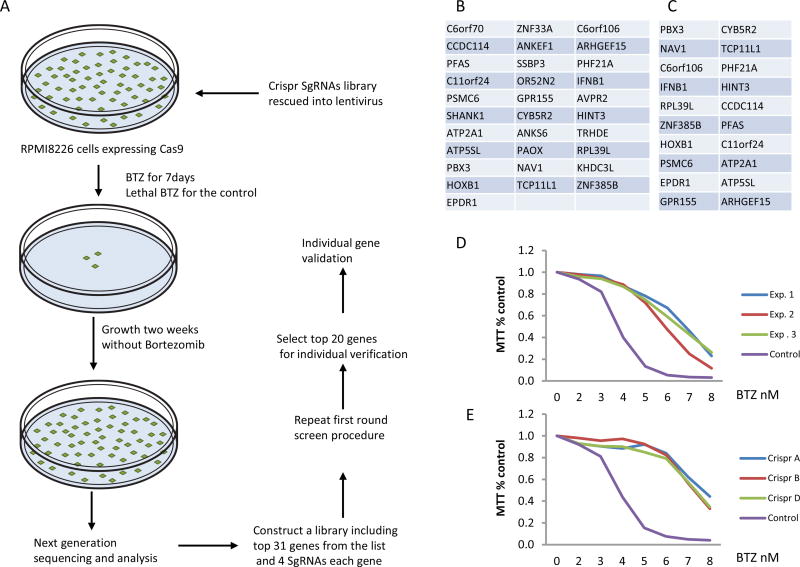
Experimental screening procedures are illustrated in figure 2A. The top 100 sgRNAs that featured in next generation sequencing of surviving cells following BTZ treatment are listed in supplemental 2. The 31 most abundant of these sgRNAs found in cells surviving BTZ treatment were selected for secondary screening (Figure 2B). Each of the 31 potential BTZ resistance inducing genes was targeted by four sgRNAs, including three sgRNAs from the GeCKO V2 library A and the fourth from GeCKO V2 library B (Supplement 3). In addition, 14 non-specific oligos with high copy number in the next generation sequencing were included in the secondary screening as controls (supplemental 3). We performed secondary screening in quadruplicate and next generation sequencing was again performed in the resistant cells. Sequencing results were listed at supplemental 4. The 20 most abundant sgRNAs (Figure. 2C) were then selected for individual validation. Four sgRNAs (the same sequence was used as in the secondary screen) for each of these 20 genes were individually cloned and mixed for virus packaging. Ultimately, only the PSMC6 gene was identified as consistently inducing BTZ resistance in independent screenings (Figure.2D). Next, we infected RPMI8226 cells with individually packaged sgRNA targeting PSMC6. Three of four sgRNA were associated with resistance to BTZ (Figure. 2E).
Figure 2.
PSMC6 was the only gene producing resistance to BTZ after knocking out by CRISPR sgRNA. (A) Schematic diagram for genome wide sgRNA screen. (B) Top gene list for first round screen. (C) Top gene list for second screen. (D) Three independent experiments (MTT assay) of RPMI8226 cells knocked out by a mix of four PSMC6 sgRNAs (A+B+C+D). (E) PSMC6 knock out by three independent sgRNAs produces resistance to bortezomib (BTZ).
PSMC6 was validated as a BTZ-sensitive gene
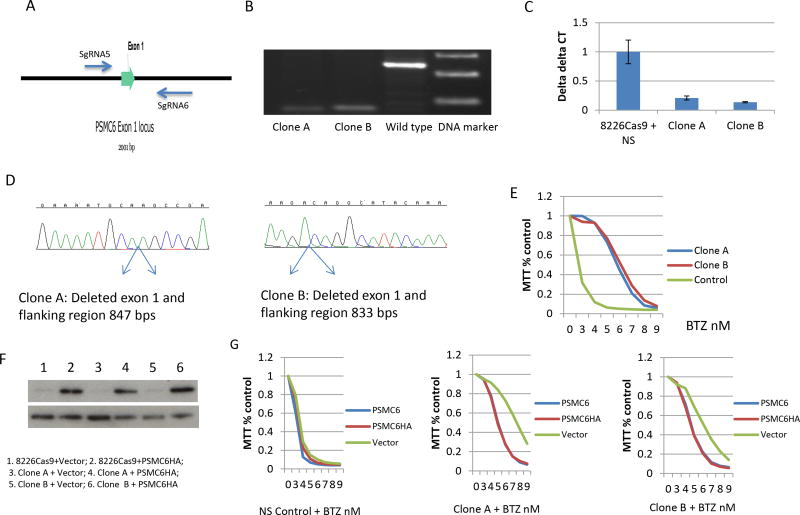
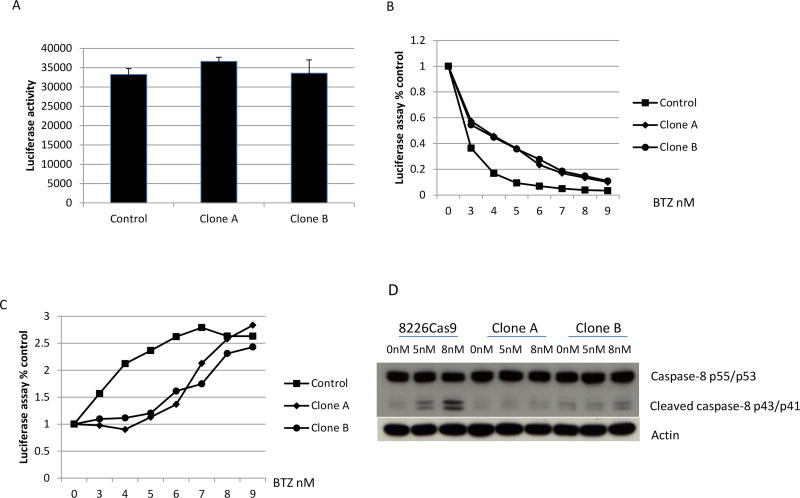
To confirm that PSMC6 deficiency correlates with BTZ resistance, we designed a pair of sgRNA (sgRNA # 5 and 6) targeting intron regions flanking exon 1 of PSMC6 (Figure 3A). This pair of sgRNA only deletes endogenous PSMC6 exon 1, but not PSMC6 cDNA introduced by a vector, thus allowing the rescue of any PSMC6 deficiency phenotype by over-expression of wild type PSMC6. These sgRNAs delete around 800 bps, allowing screening for PSMC6 knockout by PCR. We subsequently obtained two clones (clones A and B) from RPMI8226 cells with PSMC6 deficiency validated by PCR, quantitative PCR and Sanger sequencing (Figure.3B-D). Both clones developed resistance to BTZ with an IC50 for clone A and B of 6.0 nm compared to parental cells in which the IC50 is 2.5 nM. (Figure. 3E). Furthermore, MM cells lacking PSMC6 also developed resistance against Carfilzomib (CAR) (Supplemental Figure 1A). These results were fully reproduced in cell line KMS11 (Supplemental Figure 1B&C).
Figure 3.
PSMC6 was validated as a BTZ-sensitive gene. (A) A pair of sgRNAs targeting intron region flanking Exon 1. (B) PSMC6 exon 1 deletion was validated by PCR for two cloned cells. (C) PSMC6 expression levels detected by quantitative PCR. (D) Sanger sequencing analyze cloned RPMI8226 cells deleted PSMC6. PCR amplified PSMC6 exon 1 region and cloned into T-vector. Then purified plasmid was analyzed by Sanger sequencing. (E) MTT assay for cloned RPMI8226 cells (clone A and B) deleted PSMC6. The cells were treated with BTZ for 72 hours. Control=RPMI8226 cells infected with lenti-vector with non-specific sgRNA. (F) Over expression of PSMC6 with HA tag in cloned PSMC6 deficiency cells. (G) Over expression of PSMC6 wild type or with HA tag rescues the phenotype of PSMC6 deficiency. NS= non-specific sgRNA control. bortezomib (BTZ). Vector=empty lentivirus vector. PSMC6HA=lentivirus expressing PSMC6 with HA tag.
To exclude off target effects, we re-introduced wild-type PSMC6 with an HA tag in the two PSMC6 gRNA targeted clones (Figure 3F). In both cases, the expression of wild-type PSMC6 rescued most of the BTZ-sensitive phenotype (Figure 3G)(IC50 for parent cell is 3.7nM, IC50 for clone A was restored from 7.6 to 5.5 nM, and IC50 for clone B was restored from 6.4 to 4.6 nM).
PSMC6 deficiency was specific for BTZ resistance
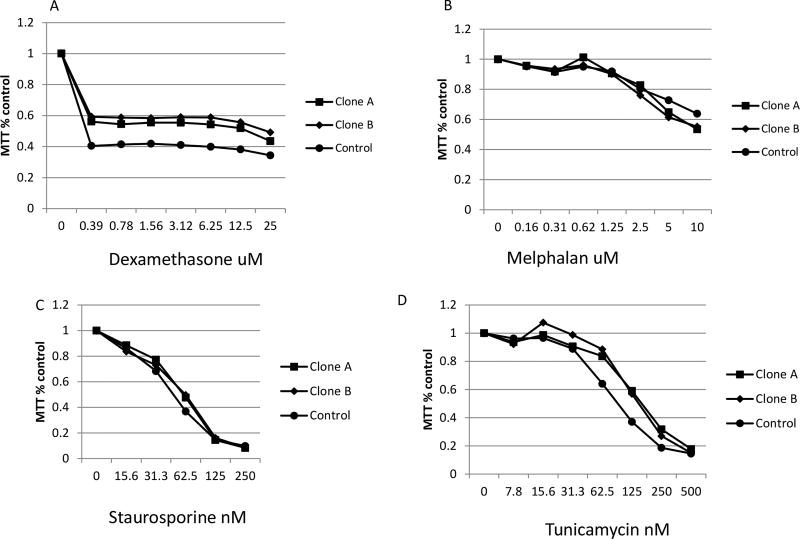
In order to confirm that drug resistance induced by PSMC6 deficiency was specific to proteasome inhibitors, we treated PSMC6 deficient RPMI8226 cells with dexamethasone, melphalan, tunicamycin (an ER stress inducer) and staurosporine (a pan kinase inhibitor). Isogenic PSMC6 wild-type and PSMC6 knockout RPMI8226 (Figure 4) or similarly engineered KMS11 cells did not show any change in sensitivity to dexamethasone (supplemental Figure 2A) and melphalan (supplemental Figure 2B).
Figure 4.
MM cells without PSMC6 have no significant difference in sensitivity to others chemicals tested. Cloned RPMI8226 deleted for PSMC6 by CRISPR sgRNA (PSMC6Cr5+6) were tested for response to different chemicals assayed by MTT (72 hours treatment). (A) Dexmethasone. (B) Melphalan. (C) Tunicamycin. (D) Staurosporine.
Mechanism of BTZ resistance induced by PSMC6 deficiency
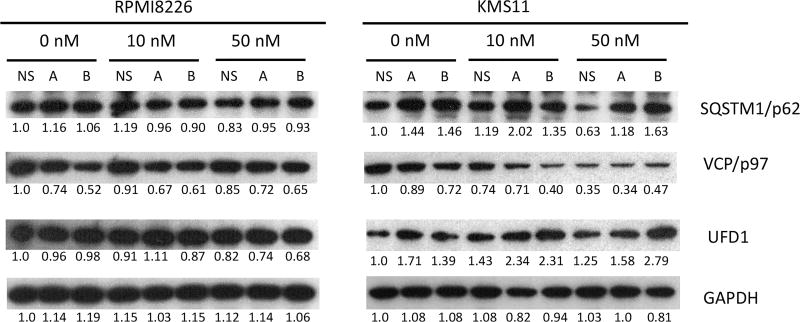
The basal level of proteasomal chymotrypsin-like activity has no significant change after deletion of PSMC6 (Figure 5A) In contrast, the proteasomal chymotrypsin-like activity in cells lacking PSMC6 was significantly reduced following exposure to BTZ (Figure 5B). MM cells with PSMC6 knockout were resistant to BTZ-induced apoptosis, which was confirmed by measuring Caspases 3/7 activity (Figure 5C) and Caspase 8 expression (Figure 5D). We checked expression levels of SQSTM1/p62, an autophagy pathway protein (35) and found no significant change in 8226 cells with or without PSMC6 protein, however the protein expression level was significantly increased in KMS11 cells after deletion of the PSMC6 gene.(Figure 6). Expression levels of the endoplasmic reticulum-associated protein degradation gene VCP/p97, (36), were reduced for both RPMI8226 and KMS11 cells after deletion PSMC6 (Figure 6). VCP/p97 expression was further inhibited in KMS11 cells with or without PSMC6 by BTZ treatment. UFD1, which forms a complex with VCP/97(36), has similar expression change as SQSTM1/p62 (Figure 6). We next measured the total 26S proteasome and demonstrate that RPMI8226 and KMS11 cells lacking PSMC6 have varying change in 26S when compared to parental cells. However, both RPMI8226 and KMS11 cells without PSMC6 continue to express significant amounts of 26S proteasome (Supplemental Figure 2C). This is important to note since other studies have demonstrated absence of the 19S proteasome to be lethal because its absence precludes formation of functional 26S proteasomes. It seems this no longer applies in the CRSIPR knockout MM cells.
Figure 5.
MM cells without PSMC6 showed resistance to bortezomib (BTZ) induced apoptosis. (A) Cell-based, chymotrypsin-like activity assay on cell with or without PSMC6 without treatment with BTZ. (B) Cell-based, chymotrypsin-like activity assay. The cells were treated with BTZ for 24 hours. (C) Caspase 3/7 activity detected by luminescent assay. The cells were treated with BTZ for 24 hours. (D) Caspase 8 p55/53 and cleaved caspase 8 p43/41 detection by Western blot. The cells were treated with BTZ for 24 hours.
Figure 6.
Expression of SQTM1/p62, VCP/p97, and UFD1 in RPMI8226 and KMS 11 cells with or without PSMC6 treated with BTZ. Cloned RPMI8226 or KMS11 cells (clone A and B) deleted PSMC6 and Control cells infected with non-specific sgRNA (NS) treated with BTZ at 10 or 50 nM for 4 hours. The proteins expression was detected by western blot and analyzed by ImageJ software (numbers below the Figure 6 blots are normalized to GAPDH and untreated control).
Depletion of PSMC subunits PSMC1, PSMC2, PSMC3, PSMC4, and PSMC5 also results in resistance to BTZ
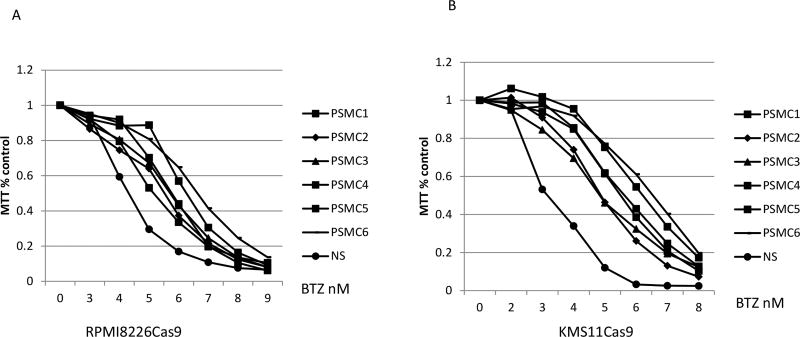
We tested the effect on BTZ resistance of the remaining PSMC subunits PSMC1 to PSMC5 using three sgRNAs for each gene, which demonstrated that each PSMC gene also caused BTZ resistance in RPMI8226 and KMS11, albeit their IC50 was less than that of PSMC6 (Figure 7A-B, IC50 were calculated by PRISM, see supplemental table S1). Cross experiment comparisons are not tested for statistical variability. The induction of resistance was confirmed by individual CRISPR sgRNAs (Supplemental Figure 2D&E). Notably, however these five subunits did not appear in our CRISPR library screening.
Figure 7.
Deficiency of PSMC1-PSMC5 also results in resistance to bortezomib (BTZ) treatment. The cells were infected with lentivirus including three sgRNAs for each gene. The infected cells were treated with BTZ at concentration of 4, 5, and 6 nM 72 hours after infection. The cells that survived to the highest concentration for drug treatment were tested for response to BTZ assayed by MTT (72 hours treatment). (A) RPMI8226 cells. (B) KMS11 cells.
PSMC6 mutation profile in Myeloma patients
We analyzed the mutation profile of all proteasome subunits in the genomic sequences derived by whole exome sequencing of 895 patients in the IA10 release of the CoMMpass study (https://www.themmrf.org/). It is important to note that the vast majority of exomes in this dataset are in newly diagnosed patients unexposed to BTZ. In that dataset a total of 36 mutations in 19S proteasome subunits were identified (one patient has three PSMC5 mutations and one patient has two PSMD7 mutations) (supplemental 5). A total of 16 PSMC subunit mutations, were identified although none of them was in PSMC6. PSMB5 is most frequently associated with BTZ resistance in MM cell lines in vitro. Two patients with PSMB5 mutations are detected in the CoMMpass dataset. Functional confirmation of the significance of these mutations was not performed.
Discussion
The mechanisms of BTZ resistance have remained elusive in MM. The proteasome subunit PSMB5 is a major target of BTZ. Mutations and increased PSMB5 expression have been correlated with BTZ resistance in vitro (8, 10, 13), but not yet in vivo (17, 18). By performing whole exome sequencing on multidrug resistant MM patients (37) we have previously identified a missense mutation in PSMG2, another component of the proteasome, as well as in XBP-1 potentially suggesting their involvement in BTZ resistance. Recently Acosta-Alvear et al. (21) using a next generation shRNA library screening demonstrated that knocking down the vast majority of subunits of the 19S proteasomal regulator, especially D group subunits PSMD2, PSMD6, PSMD12 and PSMD13 induces BTZ resistance. In their validation screening PSMC1 and 6 also appeared in the list of genes which can induce resistance.
In the current study we applied a newly developed unbiased CRISPR genome-wide screening to identify potential gene impairment correlated with BTZ resistance. After performing two independent screenings and further validation, we identified the proteasome regulatory subunit PSMC6 as the most prominent gene associated with BTZ resistant. We further demonstrated that impairment of the remaining PSMC family members also caused BTZ resistance, albeit less potently than with PSMC6. Of note, the PI resistance we observed was not complete, however the same effect was found in cells treated with BTZ and CAR, suggesting that both drugs are affected by PSMC6. Further validating our finding PSMC6 was also listed in an independent CRISPR screening performed by Sheffer et al. using the same library, although no further validations were reported (23).
PSMC6 and other PSMC family member are located in the base region of the proteasome 19S regulatory particle. All PSMC subunits have an ATPase function, which is necessary to unfold the substrates and translocate them into the core particle (38). PSMB5 is a catalytic subunit, which has chymotrypsin-like activity and is part of the proteasome 20S core particle. Our results show that PSMC6 depletion reduces the BTZ-induced chymotrypsin-like activity of PSMB5.
Acosta-Alvear et al. also performed whole genome screening using shRNA and selected PSMD12 from their top hits for validation(21). However, they found that knockdown PSMD12 did not affect the activity of the 20S subunits targeted by carfilzomib. They demonstrated that the mechanism of PSMD12 was the selective accumulation of protective factors, including SQSTM1/p62, and two subunits of a heterotrimeric complex functioning in endoplasmic reticulum-associated protein degradation UFD1L and the triple AAA ATPase VCP/p97. We therefore checked expression levels of SQSTM1/p62, VCP/p97 and UFD1 and also found significant changes in KMS11 cells deleted of PSMC6 for SQSTM1/p62 and UFD but could not demonstrate such findings in RPMI8266 cells. Since our primary screen and validation were performed on 8226 cells, the autophagy pathway seems not to be involved in BTZ resistant MM cells lacking PSMC6. In our experiments, expression levels of VCP/p97 were reduced for both RPMI8226 and KMS11 cells deleted PSMC6, a finding in contrast to the report by Acosta-Alvear et al. regarding PSMD12 (21). Franke et al. (10) found that BTZ binding to mutated PSMB5 was impaired in BTZ resistant MM cells, underlying the basis for BTZ resistance in this situation. Since mutations of PSMC6 have not yet been identified one possible explanation of our finding is that PSMC6 ablation changes 26 proteasome structure and influences bortezomib binding to PSMB5. However this needs to be addressed by crystal structure studies.
Using another approach (gene-trap) Tsvetkov et al. have reached a similar conclusion, to our work, demonstrating that a modest reduction in PSMC2 - 6 protected cells from proteasome inhibitors. (22) One of their findings was that strong 19S reduction was cytotoxic, for they were unable to isolate stable clones deleted of the 19S subunit from the resistant pools of cells, unable to recover diploid cell variants depletion of PSMD12 by CRISPR technology and unable to recover clones with inactivated PSMC2 and PSMD12 from haploid mouse embryonic stem cells that harbor reversible gene-trap induced by Cre-mediated inversion. However in our study we developed both RPMI8226 and KMS11 cells with homozygous deletion of PSMC6 successfully. The knockout cells were healthy under microscopy. and expressed 26S proteasomes. One possible explanation is that PSMC6 function is cell dependent, since Tsvetkov et al, using haploid and near-haploid cell lines (near-haploid human chronic myeloid leukemia cells (KBM7), near-haploid fibroblast cells (HAP1) and haploid mouse embryonic stem cells). (22) Whether other 19S proteasome subunits are also dispensable in MM cells needs further study in future.
Genome-wide sequencing data of PI resistant MM are currently not available. We did however analyze the 895 patients sequenced as part of the MMRF CoMMpass trial and found only one mutation in PSMC6, however a significant number of patients with other 19S subunits mutations (3.7%) are noted. Most of these patients are newly diagnosed and we hypothesize that mutations would be enriched in resistant/refractory cases to PIs, in line with our recent findings in patients that have relapsed after treatment with IMiDs (19). Thus, CRBN, the gene targeted by IMiDs, was found mutated in only 0.2% of newly diagnosed MM (https://research.themmrf.org/rp/mycommpass), but in 12% of patients refractory to IMiDs therapies with mutations in the CRBN pathway in at least 22% of the same patients and probably higher since structural variations were not analyzed (19). Acosta-Alvear et al. (21) found that PSMC2 expression levels quantified by flow cytometry were significantly higher in the group of patients who achieved complete response than partial responders after treatment with carfilzomib-based therapy. A similar longitudinal and comprehensive screening is desired in patients treated with PIs in order to elucidate the mutation rate and expression levels of PSMC genes over time and their potential role in the development of PI resistance in human MM.
In summary, using comprehensive genome wide CRISPR screening we identified and validated PSMC6 depletion as the strongest hit associated with BTZ resistance. PSMC 1–5 subunits also confer BTZ resistance in MM cells. PSMC6 deficiency reduced the ability of BTZ to suppress the chymotrypsin-like activity of PSMB5. Three independent studies with different approaches have now reached the same conclusion that deletion or down regulation of the 19S proteasome subunits leads to resistance to 20S proteasome inhibitors.
Supplementary Material
Acknowledgments
A.K.S is supported by National Institute of Health (grant RO1CA183968-03 and RO1 167511-04)
References
- 1.Shah JJ, Orlowski RZ. Proteasome inhibitors in the treatment of multiple myeloma. Leukemia. 2009;23:1964–79. doi: 10.1038/leu.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moreau P, Richardson PG, Cavo M, Orlowski RZ, San Miguel JF, Palumbo A, et al. Proteasome inhibitors in multiple myeloma: 10 years later. Blood. 2012;120:947–59. doi: 10.1182/blood-2012-04-403733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laubach JP, Schlossman RL, Mitsiades CS, Anderson KC, Richardson PG. Thalidomide, lenalidomide and bortezomib in the management of newly diagnosed multiple myeloma. Expert Rev Hematol. 2011;4:51–60. doi: 10.1586/ehm.10.83. [DOI] [PubMed] [Google Scholar]
- 4.Rosenthal A, Kumar S, Hofmeister C, Laubach J, Vij R, Dueck A, et al. A Phase Ib Study of the combination of the Aurora Kinase Inhibitor Alisertib (MLN8237) and Bortezomib in Relapsed Multiple Myeloma. Br J Haematol. 2015 doi: 10.1111/bjh.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar SK, Berdeja JG, Niesvizky R, Lonial S, Laubach JP, Hamadani M, et al. Safety and tolerability of ixazomib, an oral proteasome inhibitor, in combination with lenalidomide and dexamethasone in patients with previously untreated multiple myeloma: an open-label phase 1/2 study. Lancet Oncol. 2014;15:1503–12. doi: 10.1016/S1470-2045(14)71125-8. [DOI] [PubMed] [Google Scholar]
- 6.Reeder CB, Reece DE, Kukreti V, Mikhael JR, Chen C, Trudel S, et al. Long-term survival with cyclophosphamide, bortezomib and dexamethasone induction therapy in patients with newly diagnosed multiple myeloma. Br J Haematol. 2014;167:563–5. doi: 10.1111/bjh.13004. [DOI] [PubMed] [Google Scholar]
- 7.Stewart AK, Richardson PG, San-Miguel JF. How I treat multiple myeloma in younger patients. Blood. 2009;114:5436–43. doi: 10.1182/blood-2009-07-204651. [DOI] [PubMed] [Google Scholar]
- 8.Niewerth D, Jansen G, Assaraf YG, Zweegman S, Kaspers GJ, Cloos J. Molecular basis of resistance to proteasome inhibitors in hematological malignancies. Drug Resist Updat. 2015;18:18–35. doi: 10.1016/j.drup.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Oerlemans R, Franke NE, Assaraf YG, Cloos J, van Zantwijk I, Berkers CR, et al. Molecular basis of bortezomib resistance: proteasome subunit beta5 (PSMB5) gene mutation and overexpression of PSMB5 protein. Blood. 2008;112:2489–99. doi: 10.1182/blood-2007-08-104950. [DOI] [PubMed] [Google Scholar]
- 10.Franke NE, Niewerth D, Assaraf YG, van Meerloo J, Vojtekova K, van Zantwijk CH, et al. Impaired bortezomib binding to mutant beta5 subunit of the proteasome is the underlying basis for bortezomib resistance in leukemia cells. Leukemia. 2012;26:757–68. doi: 10.1038/leu.2011.256. [DOI] [PubMed] [Google Scholar]
- 11.Ri M, Iida S, Nakashima T, Miyazaki H, Mori F, Ito A, et al. Bortezomib-resistant myeloma cell lines: a role for mutated PSMB5 in preventing the accumulation of unfolded proteins and fatal ER stress. Leukemia. 2010;24:1506–12. doi: 10.1038/leu.2010.137. [DOI] [PubMed] [Google Scholar]
- 12.Balsas P, Galan-Malo P, Marzo I, Naval J. Bortezomib resistance in a myeloma cell line is associated to PSMbeta5 overexpression and polyploidy. Leuk Res. 2012;36:212–8. doi: 10.1016/j.leukres.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Ruckrich T, Kraus M, Gogel J, Beck A, Ovaa H, Verdoes M, et al. Characterization of the ubiquitin-proteasome system in bortezomib-adapted cells. Leukemia. 2009;23:1098–105. doi: 10.1038/leu.2009.8. [DOI] [PubMed] [Google Scholar]
- 14.Leung-Hagesteijn C, Erdmann N, Cheung G, Keats JJ, Stewart AK, Reece DE, et al. Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell. 2013;24:289–304. doi: 10.1016/j.ccr.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ling SC, Lau EK, Al-Shabeeb A, Nikolic A, Catalano A, Iland H, et al. Response of myeloma to the proteasome inhibitor bortezomib is correlated with the unfolded protein response regulator XBP-1. Haematologica. 2012;97:64–72. doi: 10.3324/haematol.2011.043331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn DJ, Berkova Z, Jones RJ, Woessner R, Bjorklund CC, Ma W, et al. Targeting the insulin-like growth factor-1 receptor to overcome bortezomib resistance in preclinical models of multiple myeloma. Blood. 2012;120:3260–70. doi: 10.1182/blood-2011-10-386789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Politou M, Karadimitris A, Terpos E, Kotsianidis I, Apperley JF, Rahemtulla A. No evidence of mutations of the PSMB5 (beta-5 subunit of proteasome) in a case of myeloma with clinical resistance to Bortezomib. Leuk Res. 2006;30:240–1. doi: 10.1016/j.leukres.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 18.Lichter DI, Danaee H, Pickard MD, Tayber O, Sintchak M, Shi H, et al. Sequence analysis of beta-subunit genes of the 20S proteasome in patients with relapsed multiple myeloma treated with bortezomib or dexamethasone. Blood. 2012;120:4513–6. doi: 10.1182/blood-2012-05-426924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kortum KM, Mai EK, Hanafiah NH, Shi CX, Zhu YX, Bruins L, et al. Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood. 2016 doi: 10.1182/blood-2016-02-698092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mimura N, Fulciniti M, Gorgun G, Tai YT, Cirstea D, Santo L, et al. Blockade of XBP1 splicing by inhibition of IRE1alpha is a promising therapeutic option in multiple myeloma. Blood. 2012;119:5772–81. doi: 10.1182/blood-2011-07-366633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Acosta-Alvear D, Cho MY, Wild T, Buchholz TJ, Lerner AG, Simakova O, et al. Paradoxical resistance of multiple myeloma to proteasome inhibitors by decreased levels of 19S proteasomal subunits. Elife. 2015;4:e08153. doi: 10.7554/eLife.08153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsvetkov P, Mendillo ML, Zhao J, Carette JE, Merrill PH, Cikes D, et al. Compromising the 19S proteasome complex protects cells from reduced flux through the proteasome. Elife. 2015;4 doi: 10.7554/eLife.08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michal Sheffer YH, Shalem Ophir, Sanjana Neville, Dhimolea Eugen, Sarkar Subhashis, Bariteau Megan A, Aftab Blake T, Groen Richard WJ, Zhang Feng, Mitsiades Constantine S. Genome-scale Crispr-Cas9 knockout studies reveal mutifactorial and functionally overlapping mechanisms of myeloma cell resistance to proteasome inhibition. ASH abstracts. 2014 [Google Scholar]
- 24.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–7. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–4. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takatsuki A, Kawamura K, Okina M, Kodama Y, Ito T, Tamura G. Structural Elucidation of Tunicamycin .2. Structure of Tunicamycin. Agr Biol Chem Tokyo. 1977;41:2307–9. [Google Scholar]
- 30.Furusaki A, Hashiba N, Matsumoto T, Hirano A, Iwai Y, Omura S. X-Ray Crystal-Structure of Staurosporine - New Alkaloid from a Streptomyces Strain. J Chem Soc Chem Comm. 1978:800–1. [Google Scholar]
- 31.Furusaki A, Hashiba N, Matsumoto T, Hirano A, Iwai Y, Omura S. The Crystal and Molecular-Structure of Staurosporine, a New Alkaloid from a Streptomyces Strain. B Chem Soc Jpn. 1982;55:3681–5. [Google Scholar]
- 32.Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–67. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 33.Jordan M, Schallhorn A, Wurm FM. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 36.Wolf DH, Stolz A. The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim Biophys Acta. 2012;1823:117–24. doi: 10.1016/j.bbamcr.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Egan JB, Kortuem KM, Kurdoglu A, Izatt T, Aldrich J, Reiman R, et al. Extramedullary myeloma whole genome sequencing reveals novel mutations in Cereblon, proteasome subunit G2 and the glucocorticoid receptor in multi drug resistant disease. Br J Haematol. 2013;161:748–51. doi: 10.1111/bjh.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattacharyya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol. 2014;15:122–33. doi: 10.1038/nrm3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.