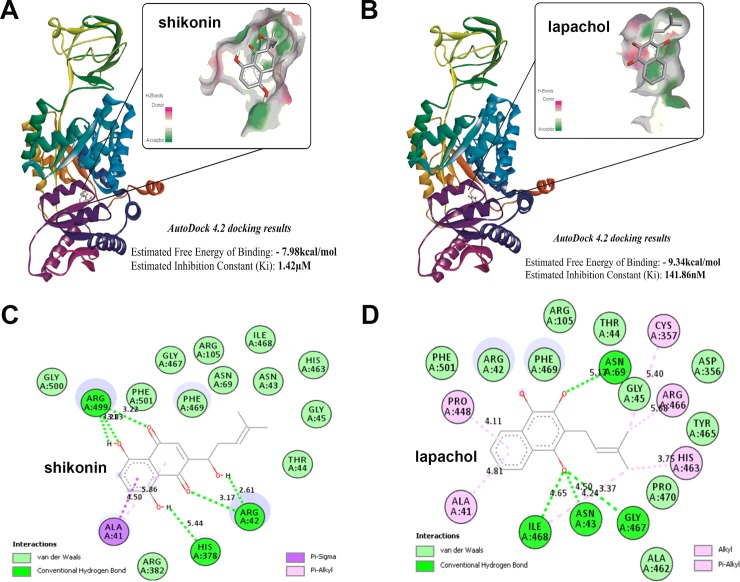
Fig 3. In silico studies of lapachol-PKM2 interactions.
(A) The structure of human muscle Pyruvate kinase M2 [1ZJH—Receptor] docked to shikonin (ligand) using Autodock 4.2 software and ligand receptor interaction diagram generated using Discovery studio 2.5. The predicted free energy of binding for shikonin -7.98 kcal/mol and predicted inhibition constant was 1.42 μM. Inset shows the site pocket with surface charge distribution on donor and acceptor atoms. (B) The structure of human Pyruvate kinase M2 docked to lapachol and ligand receptor interaction diagram generated using Discovery studio 2.5. Inset shows the site pocket with surface charge distribution on donor and acceptor atoms. (C) The 17 amino acid residues of human PKM2 protein predicted to interact with the functional groups in shikonin and the type of interaction Van der waals (pale green), H-bond (bright green), Pi-Sigma (purple) and Pi-Alkyl (pink) are highlighted. (D) The 20 amino acid residues of human PKM2 protein predicted to interact with the functional groups in lapachol and the type interaction are highlighted.