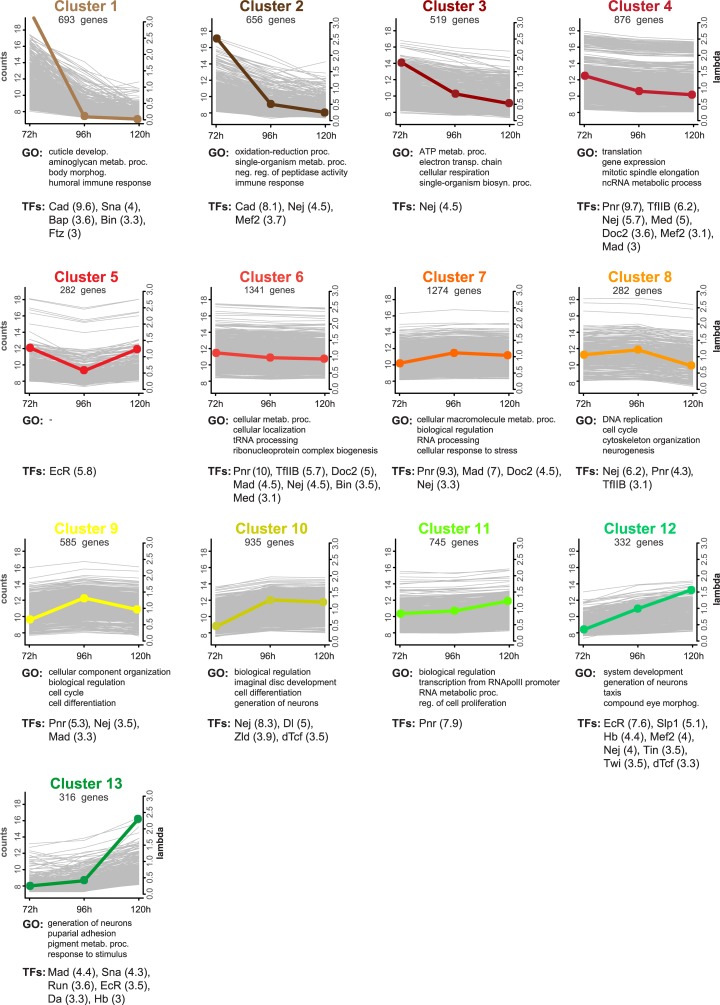
Fig 2. Co-expression clusters.
All 13 profiles of co-expressed genes predicted by HTSCluster (see Materials and Methods). The number of genes assigned to a particular cluster are indicated below the cluster name. Colored dots represent relative expression levels (lambda value) of the genes of that cluster (y-axis on the right) at each stage. Background grey lines represent the normalized mean count of all genes belonging to a cluster (y-axis on the left). Below each cluster plot, the first four non-redundant GO terms enriched in the genes of that cluster are listed (see S1 Table) and the significantly enriched transcription factors (NES > 3, see S2 Table) are shown (the NES value is given in brackets).