Abstract
Pseudomonas chlororaphis 30-84 is a biological control agent selected for its ability to suppress diseases caused by fungal pathogens. P. chlororaphis 30-84 produces three phenazines: phenazine-1-carboxylic acid (PCA), 2-hydroxy-phenazine-1-carboxylic acid (2OHPCA) and a small amount of 2-hydroxy-phenazine (2OHPHZ), and these are required for fungal pathogen inhibition and wheat rhizosphere competence. The two, 2-hydroxy derivatives are produced from PCA via the activity of a phenazine-modifying enzyme encoded by phzO. In addition to the seven biosynthetic genes responsible for the production of PCA, many other Pseudomonas strains possess one or more modifying genes, which encode enzymes that act independently or together to convert PCA into other phenazine derivatives. In order to understand the fitness effects of producing different phenazines, we constructed isogenic derivatives of P. chlororaphis 30-84 that differed only in the type of phenazines produced. Altering the type of phenazines produced by P. chlororaphis 30-84 enhanced the spectrum of fungal pathogens inhibited and altered the degree of take-all disease suppression. These strains also differed in their ability to promote extracellular DNA release, which may contribute to the observed differences in the amount of biofilm produced. All derivatives were equally important for survival over repeated plant/harvest cycles, indicating that the type of phenazines produced is less important for persistence in the wheat rhizosphere than whether or not cells produce phenazines. These findings provide a better understanding of the effects of different phenazines on functions important for biological control activity with implications for applications that rely on introduced or native phenazine producing populations.
Keywords: biological control, eDNA, phenazine, Pseudomonas chlororaphis 30-84, biofilm
Most plant-beneficial pseudomonads selected for their ability to suppress plant diseases produce a variety of secondary metabolites that are inhibitory to pathogens (Haas and Défago, 2005). Phenazines, a class of diffusible, heterocyclic compounds, are among the well-studied secondary metabolites shown to be important for biological control (Mavrodi et al., 2006; Pierson and Thomashow, 1992). Different Pseudomonas strains often produce one or more phenazines, derived from the substitution of various functional groups on the ring structure of the core phenazine, phenazine-1-carboxylic acid (PCA). PCA synthesis is mediated by a highly conserved phenazine biosynthetic operon phzXYFABCD (P. chlororaphis 30-84 nomenclature, corresponding to phzABCDEFG using P. aeruginosa annotation) (Mavrodi et al., 2001; Pierson et al., 1995). In most phenazine producing pseudomonads, the phenazine biosynthetic operon is flanked by one or more genes that encode the enzymes responsible for converting PCA to the different phenazine derivatives either by methylation, transamination, hydroxylation, or decarboxylation (Chin-A-Woeng et al., 2001b; Delaney et al., 2001; Mavrodi et al., 2001, 2006). For example, the genome of the biological control strain P. chlororaphis 30-84 contains the modifying gene phzO, which encodes a monooxygenase responsible for the conversion of some PCA to 2OHPCA (Delaney et al., 2001; Pierson and Pierson, 2010). The genome of the opportunistic human pathogen P. aeruginosa PAO1 has several phenazine modifying genes that convert PCA into four additional phenazine derivatives: phenazine-1-carboxamide (PCN), 5-methyl-phenazine-1-carboxylic acid (5MPCA), 1-hydroxy-phenazine (1OHPZ) via the activity of phzH, phzM, and phzS, respectively, and pyocyanin (PYO) via the activity of both phzM and phzS (Mavrodi et al., 2001, 2006).
As a group, phenazine compounds are electro-chemically active metabolites that contribute to bacterial fitness in part via antagonistic effects on competitors or eukaryotic host cells through the generation of reactive oxygen species (ROS) including superoxide (O2−) and hydrogen peroxide (H2O2) (Mavrodi et al., 2001; Pierson and Pierson, 2010; Price-Whelan et al., 2006). Once phenazines exit the producing-cells, they may become incorporated into the membranes of other organisms, resulting in the uncoupling of oxidative phosphorylation, the generation of ROS, and ultimately interference in electron flow and enzyme functions related to cellular respiration (Baron et al., 1989; Gu and Imlay, 2011). Phenazines are associated with the virulence of the opportunistic in human pathogen P. aeruginosa due to phenazine-induced changes in the redox balance of host cells, resulting in injury to the host cells or imbalanced oxidative stress responses (Price-Whelan et al., 2006). Phenazines produced by plant-beneficial pseudomonads are associated with their ability to inhibit the growth of a diversity of eukaryotic plant pathogens, including fungi and nematodes (Cezairliyan et al., 2013; Chin-A-Woeng et al., 2001a; Mavrodi et al., 2006; Pierson and Pierson, 2010; Zhou et al., 2016) and play a role in the competitive survival of producers in the rhizosphere (Chin-A-Woeng et al., 2003; Mazzola et al., 1992).
Phenazines also contribute to host fitness via their roles in biofilm formation and conditioning of the biofilm environment. The essential role of phenazines for biofilm formation was demonstrated in studies that showed phenazine-defective mutants were significantly impaired in cell attachment and biofilm formation, compared to wild-type (Das et al., 2015; Maddula et al., 2006, 2008; Ramos et al., 2010; Selin et al., 2009; Wang et al., 2011). Similarly, phenazines were found to be important for the formation of unattached, floating biofilms (Wang et al., 2016). Altering the ratio of the two major phenazine derivatives in P. chlororaphis 30-84 also affected biofilm development and architecture (Maddula et al., 2008).
Phenazines have been linked to the production of bacterial exopolymeric substances (EPS), structural components of the extracellular matrix that cells produce during biofilm development (Das et al., 2013a, 2013b). This EPS matrix protects bacterial cells from physical and mechanical stresses (i.e., desiccation, antibiotics, host immune system), having a major role in their survival and persistence in natural environments, including plant-associated niches (Ghosh and Maiti, 2016; Gunn et al., 2016; Jayathilake et al., 2017; Okshevsky and Meyer, 2015; Whitchurch et al., 2002). The composition of the extracellular matrix may vary between strains and is complex, including lipopolysaccharides, glycolipids, lipids, proteins or peptides, nucleic acids, and certain secondary metabolites [reviewed in (Das et al., 2013b; Flemming and Wingender, 2010; Mann and Wozniak, 2012; Steinberg and Kolodkin-Gal, 2015; Wei and Ma, 2013)]. Recent attention has focused on extracellular DNA (eDNA), found to be a ubiquitous component of the biofilm matrix, with hypothesized roles relating to initial bacterial adhesion and surface aggregation (Das et al., 2010), biofilm development (Gloag et al., 2013), promoting bacterial binding affinities with polymers and metabolites (Das et al., 2013b), and development of cation gradients for protection against antibiotics (Mulcahy et al., 2008). Phenazine production plays a role in eDNA release, and subsequently in the structure of the biofilm matrix produced. For example, the generation of hydrogen peroxide concomitant with PYO production by P. aeruginosa is required for eDNA release and biofilm production (Das and Manefield, 2012). Similarly, in P. chlororaphis 30-84 phenazine production stimulated the release of eDNA and was found to be vital to the production of the structured biofilm matrix (Wang et al., 2016). In the latter study, the production of structured matrix was abolished if eDNA was degraded by DNase I. It was postulated that eDNA release and biofilm matrix production are bacterial mechanisms for ameliorating ROS stress associated with phenazine production (Das and Manefield, 2012).
From a biological control perspective, the type of phenazines produced may affect the capacity of the producing organism to suppress disease at many levels, including the ability to inhibit different pathogens, form biofilms, and competitively colonize and survive in the rhizosphere microbiome. Despite the importance of phenazines for biological control activity, it is not clear how the type of phenazine produced affects any of these important capacities. Previous studies comparing biological control strains that produce different phenazines revealed differences in the spectrum of pathogen inhibition (Chin-A-Woeng et al., 2003; Mavrodi et al., 2006). Moreover, because the biological control strains studied were isolated from different plant hosts, it could be speculated that specific phenazines may play a role in bacterial recruitment to and survival in the rhizospheres of different hosts. Differences between phenazines in their functional groups also likely change their solubility and capacity for electron shuttling, especially at different pH levels (Cezairliyan et al., 2013; Chin-A-Woeng et al., 2003; Pierson and Pierson, 2010). In most previous studies, understanding the function of different phenazines has been complicated by their being produced by different bacterial strains.
In the present study, in order to more directly investigate the effects of the production of different phenazines on biological control activity, isogenic lines producing PCN, 1OHPZ, 5MPCA, or PYO in addition to the core phenazine PCA were created. P. chlororaphis 30-84, a biological control strain selected for its efficacy in suppressing take-all disease of wheat caused by the ascomycete pathogen Gaeumannomyces graminis var. tritici (Ggt), was used as the target biological control strain for this purpose (Pierson and Thomashow, 1992). Previous work demonstrated a critical role for phenazine production in Ggt inhibition, disease suppression, and rhizosphere competence (Mazzola et al., 1992; Pierson and Thomashow, 1992). The new, altered phenazine producers, together with the previously characterized strains P. chlororaphis 30-84 wild-type (30-84WT), a phenazine-deficient mutant (30-84ZN), and 30-84PCA (Maddula et al., 2008; Wood et al., 1997), were used to study the effect of the type of phenazines produced on biological control activity, including pathogen inhibition, take-all disease suppression, biofilm formation, eDNA release and wheat root colonization.
Materials and Methods
Bacteria strains and growth condition
All bacterial strains and plasmids used in this study are described in Table 1, and primers are listed in Table 2. Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) medium. Pseudomonas chlororaphis strain 30-84 and its derivatives were grown at 28°C in either LB medium containing 5 g of NaCl per liter, pigment production medium (PPMD) and AB minimal medium supplemented with 2% casamino acids (Difco, Franklin Lakes, NJ, USA) (AB-C). Antibiotics were used where appropriate at the following concentration for E. coli: ampicillin (Ap) at 100 μg/ml, gentamicin (Gm) 15 μg/ml, Kanamycin (Km) at 50 μg/ml and tetracycline (Tc) at 25 μg/ml; for P. chlororaphis: Gm at 50 μg/ml, rifampicin (Rif) at 100 μg/ml, Km at 50 μg/ml, and Tc at 50 μg/ml.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| P. chlororaphis | ||
| 30-84WT | PCA+, 2-OH-PCA+, 2-OH-PHZ+, RifR, Wild type | Pierson and Thomashow (1992) |
| 30-84PCA | PCA+, phzO::Tn5, KmR | Maddula et al. (2008) |
| 30-84O* | PCA+, 2-OH-PCA+, 2-OH-PHZ+, RifR, 2OHPCA over producer, 30-84WT containing plasmid pKM-phzO* | Maddula et al. (2008) |
| 30-84ZN | Phz−, phzB::lacZ | Wood et al. (1997) |
| 30-84H | PCA+, PCN+, 30-84PCA containing plasmid pGT2-phzH | This study |
| 30-84S | PCA+, 1OHPZ+, 30-84PCA containing plasmid pGT2-phzS | This study |
| 30-84M | PCA+, 5MPCA+, 30-84PCA containing plasmid pGT2-phzM | This study |
| 30-84MS | PCA+, PYO+, 30-84PCA containing plasmid pGT2-phzMS | This study |
| ATCC17411 | PCA+, PCN+ | Mavrodi et al. (2001) |
| P. synxantha 2-79 | PCA+ | Weller (1983) |
| E. coli | ||
| DH5α | F-recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 Δ(argF-lacZYA)II69 ϕ80lacZ ΔM15 λ- | Gibco-ThermoFisher (Waltham, MA, USA) |
| HB101 | F- hsdS20(rB-mB-) supE44 recA1 ara14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-5 | Gibco-ThermoFisher |
| Plasmid | ||
| pCR2.1-TOPO | AmpR, KmR, T7 promoters, f1 ori, lacZα | Invitrogen Life Technologies |
| TOPO-phzH | pCR2.1-TOPO carrying a 1.9-kb DNA fragment containing phzH gene | This study |
| TOPO-phzS | pCR2.1-TOPO carrying a 1.2-kb DNA fragment containing phzS gene | This study |
| TOPO-phzM | pCR2.1-TOPO carrying a 1.2-kb DNA fragment containing phzM gene | This study |
| TOPO-phzM_BB | pCR2.1-TOPO carrying a 1.2-kb DNA fragment containing phzM gene | This study |
| pGT2 | pProbe-GT′: pVS1 replicon, p15a origin of replication, gfp transcriptional fusion; GmR | Miller et al. (2000) |
| pGT2-PPsP | pGT2 carrying a 127-bp DNA fragment containing truncated promoter sequence of phzX gene (PPsP) | This study |
| pKT2- PPsPlacZ | pKT2 carrying a 127-bp DNA fragment containing truncated promoter sequence of phzX gene (PPsP) fused to lacZ; KmR | Yu (2016) |
| pGT2-phzH | pGT2-PsP carrying a 1.9-kb DNA fragment containing phzH gene | This study |
| pGT2-phzS | pGT2-PsP carrying a 1.2-kb DNA fragment containing phzS gene | This study |
| pGT2-phzM | pGT2-PsP carrying a 1.2-kb DNA fragment containing phzM gene | This study |
| pGT2-phzMS | pGT2-PsP carrying a 2.4-kb DNA fragment containing phzM and phzS gene together | This study |
KmR, AmpR, GmR and RifR = kanamycin, ampicillin, gentamicin and rifampin resistance, respectively.
Table 2.
Oligonucleotides used for gene cloning
| Primers | Sequences (5′-3′)a |
|---|---|
| phzH1 | CGCACGGATCCTTTCAGCATGTTC (BamHI) |
| phzH2 | CGCACAAGCTTCGTCACGCTCA (HindIII) |
| phzM1 | CGCACGGATCCTTTCGGTACGCAGGAAAAG (BamHI) |
| phzM2 | CGCACAAGCTTGTTGAAAGTTCCGATTCA (HindIII) |
| phzM3 | CGCACGGATCCGTTGAAAGTTCCGATTCA (BamHI) |
| phzS1 | CGCACGGATCCAAAAGGAAGCACC (BamHI) |
| phzS2 | CGCACAAGCTTCTAGCGTGGCCGTT (HindIII) |
Underlined nucleotides are restriction sites added and the restriction enzymes are indicated at the end of primers.
DNA manipulations and sequence analysis
Standard methods were used for plasmid DNA isolation, transformation, cloning, restriction enzyme digestions, agarose gel electrophoresis, and T4 DNA ligation (Sambrook and Russell, 2001). Plasmids were introduced into P. chlororaphis 30-84 and its derivatives using triparental mating as previously described (Pierson and Thomashow, 1992; Yu et al., 2017) Polymerase chain reaction (PCR) was carried out using Taq DNA polymerase (New England BioLabs, Ipswich, MA, USA). DNA sequencing was performed at the Laboratory for Genome Technology within the Institute for Plant Genomics and Biotechnology, Texas A&M University, College Station, TX, USA. Nucleotide and amino acid sequences were obtained from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/) or the Pseudomonas Genome Database (http://www.pseudomonas.com/).
Construction of the altered phenazine expression vectors
The open reading frames with their native ribosome binding site sequences for phzH, phzM, and phzS from P. aeruginosa PAO1 were used to design primers to amplify each of the gene inserts. Primer pairs phzH1-phzH2, phzM1-phzM2, and phzS1-phzS2 were used to amplify the gene inserts of phzH, phzM, and phzS genes, respectively (Table 2). Following PCR, each amplicon was cloned separately into the plasmid pCR2.1-TOPO (Invitrogen Life Technologies, Carlsbad, CA, USA) by TA cloning. TOPO-phzH, TOPO-phzS, TOPO-phzM were then digested with BamHI-HindIII, and the inserts were cloned into the BamHI-HindIII site in pGT2-PPsP (Table 1), resulting in pGT-phzH, pGT2-phzS and pGT2-phzM (Table 1). To construct the plasmid pGT2-PPsP, a modified phenazine promoter of 30-84 (PPsP) was digested with EcoRI and BamHI from pKT2- PPsPlacZ (Table 1), and the 127 bp fragment was inserted into the EcoRI and BamHI sites in pGT2 (Table 1). Plasmid pKT2- PPsPlacZ contains a truncated P. chlororaphis 30-84 phenazine promoter (PPsP) with 5-fold higher expression under the control of the quorum sensing system PhzR/PhzI (Yu, 2016). In order to create a plasmid containing phzM and phzS together, primer pair phzM1-phzM3 was used to create a phzM amplicon with BamHI restriction enzyme sites at both ends of the PCR product. This amplicon was cloned into the BamHI site in pGT2-phzS resulting in pGT2-phzMS (Table 1). The direction of the inserted gene was confirmed via enzyme digestion, PCR amplification and DNA sequencing. Altered phenazine-producing strains were created by individually introducing the plasmids containing the different modifying genes in trans into 30-84PCA, which contains a chromosomal disruption of phzO (phzO::Tn5) and is capable of producing only PCA (Table 1). The vector pGT2PPsP also was introduced into 30-84WT, 30-84ZN, and 30-84PCA as empty vector controls.
Phenazine extraction
Phenazines were extracted from 30-84WT, 30-84PCA and the altered phenazine-producing strains using HyperSep C18 SPE columns (Thermo Scientific, Waltham, MA, USA). Briefly, cultures were grown for two days in 10 ml PPMD amended with appropriate antibiotics. Cell-free supernatant was prepared by centrifugation (2,600 × g) of bacterial cultures for 15 min and filter sterilization of the resulting supernatant using a 0.22 μm filter (VWR, Radnor, PA, USA). The SPE column was washed with acetonitrile and equilibrated with sterilized water three times before the cell-free supernatants were added. Phenazines were retained on the columns, which were washed three times with sterilized water. Phenazines were eluted from the columns with 1 ml of acetonitrile.
LC-MS analyses of phenazine compounds
LC-MS analysis was performed on a Surveyor HPLC system (Thermo Finnigan, San Jose, CA, USA) interfaced with quadruple ion trap mass spectrometer (LCQ-DECA, Thermo Finnigan) in the Laboratory for Biological Mass Spectrometry at Texas A&M University. The chromatographic separation was performed on Aquasil C18 column (Thermo Hypersil-Keyston, Bellefonte, PA, USA) (2.1 × 150 mm, 3 μm) with solvent A (water) and B (acetonitrile) both containing 1 mM acetic acid as mobile phase. The flow rate was set at 200 μL/min and the elution gradient was performed as follows: from 0–5 min solvent B was held at 5%, from 5–10 increased to 50% B; from 10–24 increased to 70% B, from 24–25 to 100% B; during 25–30% 100% B is maintained, from 30–31 solvent B was reduced to 5% and maintained for 4 min. Electrospray ionization (ESI) in positive ion mode was carried out by applying 4.5 kV at the spray needle to create positive ions. Sheath gas and auxiliary gas flow rates were 50 and 10 arbitrary units, respectively. Transfer capillary temperature was held at 250°C.
Pathogen inhibition in vitro assay
The ability of all strains to inhibit a variety of fungal pathogens was determined using in vitro dual culture assays. Fungal strains were obtained from the Microbiology teaching laboratory stock collection at Department of Plant Pathology and Microbiology, Texas A&M University. Fungal strains were initially cultured on potato dextrose agar (PDA) (Difco Laboratories, Detroit, MI, USA). After 5–7 days inoculation, a 3-mm-diameter plug was taken from the edge of the mycelial mats and transferred to the center of Waksman agar (WKA) (Berg et al., 2001). WKA was prepared by adding 5 g protease-peptone, 10 g glucose, 3 g beef extract, 5 g NaCl, and 20 g agar to 1 l distilled water, and adjusting its pH to 6.8. The fungal pathogens were inoculated 2 days prior to the inoculation of bacteria except for the rapidly growing strain Pythium ultimum. Bacterial strains were prepared from overnight culture grown in LB medium at 28°C with rapid agitation, adjusted to an OD620 of 0.8 and spotted at the edge of the plates (3.5 cm apart from the center). To determine antifungal activity to P. ultimum, bacteria were spotted 2 days before fungal inoculation. After 5–7 days, zones of inhibition were measured as the distance between the edge of the bacterial colony and the fungal mycelium.
Take-all suppression assay
Strains 30-84WT, 30-84PCA, and the altered phenazine producers were assessed for their ability to suppress take-all disease on wheat seedlings under growth chamber conditions as described previously (Pierson and Thomashow, 1992; Wilkinson et al., 1985). Briefly, bacterial inoculum was washed, resuspended in methyl cellulose and applied to the wheat seeds (cv. TAM112). Treated seeds were sown in tubes (25 × 200 mm) filled with a layer (5 g) of autoclaved vermiculate overlaid with 20 g of an autoclaved wheat rhizosphere soil mix (soil: sand, 2:1, v:v) amended with Gaeumannomyces graminis var. tritici (Ggt) (as colonized oat kernels fragments 0.85%, w/w). For the fungal control, autoclaved oat kernels were ground and amended to the soil mix (0.85%, w/w); for the inoculum control, seeds treated only with methyl cellulose were used. Tubes were arranged in a complete randomized block design, incubated 3 days at room temperature for germination, and transferred to a growth chamber (16°C, 12 h dark-light cycle). After 20 days, root disease was evaluated on a scale of 0–5, where 0 = no disease and 5 = nearly dead as described previously (Pierson and Thomashow, 1992).
Biofilm and extracellular DNA (eDNA) quantification
The effect of the different phenazine derivatives on biofilm formation was investigated using the crystal violet assay as previous described (Maddula et al., 2006; O’Toole and Kolter, 1998). Briefly, overnight cultures grown in AB-C medium were adjusted to an OD620 of 0.8 with fresh medium (AB-C). Each strain (1.5 μl) was re-inoculated into 150 μl of AB-C broth in triplicate wells of 96-well polystyrene plates. For the negative control, AB-C with no inoculum was used. Plates were incubated at 28°C without agitation. After 48 and 72 h incubation cell densities determined (OD620) and then the liquid was removed by inversion of the plate, followed by vigorous tapping on absorbent paper. The remaining adherent bacteria were fixed to the plates for 20 min at 50°C and then stained for 1 min with 0.1% crystal violet. Excess stain was removed, plates were washed twice with distilled water, and adherent cells were decolorized for 5 min with a 20:80 acetone:ethanol solution. A sample of each well was transferred to another 96-well plate and the surface attached cells were quantified by measuring OD540 as described previously (Maddula et al., 2006).
The quantification of floating, non-attached biofilm matrix and extracellular DNA (eDNA) was performed as previously described (Wang et al., 2016). Briefly, cells were grown under the same conditions as described above except for using 1.5 ml of cultures in 24-well polystyrene plates. To quantify the biofilm matrix, entire cultures were transferred to 2 ml tubes and centrifuged at 12,000 × g for 5 min. The supernatant was removed, and the mixture of hydrated biofilm matrix and encapsulated cells was quantified by weight. To determine the concentration of eDNA, the entire static culture was transferred to 50 ml centrifuge tubes and vortexed vigorously to thoroughly disrupt the biofilm matrix. The supernatants were collected by centrifugation (1,250 × g for 10 min) and the remaining bacterial cells were removed by passing the supernatant through a 0.22 μm filter (VWR). The concentration of eDNA was quantified using a Qubit 2.0 Fluorometer (Invitrogen Life Technologies) according to the manufacturer’s protocol.
Exogenous phenazine treatment
Overnight cultures of 30-84WT, 30-84ZN and the altered phenazine-producing strains were inoculated into 20 ml PPMD medium and grown cultures at 28°C with agitation until an OD620 of 2.4 was reached. Cell-free supernatant was collected by centrifuging bacterial cultures (1,230 × g, 20 min) and filter sterilizing the supernatant using a 0.22 μm filter (VWR). To remove eDNA residues, 30 units of DNase-I (Qiagen, Hilden, Germany) were added to the supernatants. After 1 h at room temperature, DNase-I treated supernatants were incubated at 65°C in a water-bath for 10 min to deactivate the DNase-I. The cell-free, DNase-I treated supernatants were diluted 1:10 in fresh AB-C. For the control, cell-free, DNase-I treated supernatant of 30-84ZN was used. For the assay, overnight cultures of 30-84ZN were adjusted to an OD620 of 0.8, and inoculated at 1:100 dilution into the media that contained supernatant from either 30-84WT, one of the altered phenazine-producing strains or 30-84ZN. Production of eDNA and biofilm matrix were determined as described above after 72 h.
Root colonization and rhizosphere persistence assays
Bacterial colonization and persistence were determined using a repeated plant/harvest cycle assay as described previously (Mazzola et al., 1992). Briefly, bacterial inoculum was prepared in 20 ml KMB broth and inoculated into natural wheat rhizosphere soil mix (soil:sand, 2:1, v:v) as initial populations of 108 cfu/g of soil for all treatments. Wheat seeds (cv. TAM112) were surface sterilized with 0.6% NaClO and germinated on moistened, sterile germination paper. After 3 days, the germinated seedlings were planted into plastic tubes (25 × 200 mm) containing the soil mix inoculated with either 30-84WT, 30-84ZN or one of the altered phenazine-producing strains. The tubes (60 tubes per treatment) were arranged in a randomized complete block design. Each plant/harvest cycle spanned 20 days, and 6 plants per treatment were harvested at the end of each plant/harvest cycle. Total bacterial populations on roots were determined by counting CFU on LB plates amended with appropriate antibiotics. For the plants not used in the CFU estimations, shoots were removed at the soil surface level and soil with remaining root systems were processed as previously described (Mazzola et al., 1992). Newly germinated wheat seedlings were re-planted into the processed soil and the plant/harvest cycle was repeated 5 times.
Statistical Analysis
All data are presented as the mean ± SEM from at least two experiments. Differences between strains were analyzed statistically using ANOVA and Fisher’s protected Least Significant Difference (LSD) test (P < 0.05).
Results and Discussion
Characterization of altered phenazine-producing strains of P. chlororaphis 30-84
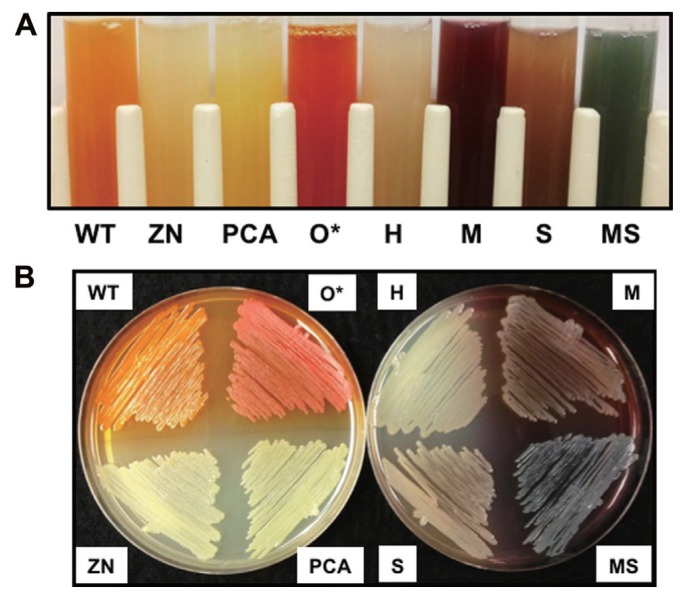
In order to examine the function of different phenazines in bacterial fitness and biological control, isogenic derivatives of P. chlororaphis 30-84 that produced different phenazines were constructed via introducing plasmids expressing each phenazine modifying gene separately into 30-84PCA (Table 1). The use of 30-84PCA (producing only PCA) rather than the wild type enabled the entire pool of PCA to be available for conversion to other phenazines and thus comparison of the different phenazine derivatives in an isogenic manner. Production of the expected phenazine by each altered phenazine-producing strain was confirmed first via colorimetric observation, because each of the different phenazines has a distinctive color at neutral pH (Haynes et al., 1956; Mavrodi et al., 2001; Pierson and Thomashow, 1992; Turner and Messenger, 1986). Compared to the bright orange phenazine produced by 30-84WT when grown to stationary phase, 30-84PCA (PCA only producer) produced a bright yellow phenazine, whereas 30-84H (PCN producer) produced a greenish-tan phenazine, 30-84M (5MPCA producer) produced a dark red phenazine, 30-84S (1OHPHZ producer) produced a tan-brown phenazine, and 30-84MS (PYO producer) produced a blue-green phenazine (Table 1, Fig. 1). Additionally, 30-84ZN (phenazine null mutant) produced no phenazines and appeared white and 30-84O* (2OHPCA overproducer) appeared dark-orange as described previously (Maddula et al., 2008) (Table 1, Fig. 1).
Fig. 1.
Production of phenazines by isogenic derivatives of P. chlororaphis 30-84. (A) Phenazine production in PPMD broth at pH 7.0 and (B) on PPMD agar plate. Strains include: P. chlororaphis 30-84 wild-type (WT), 30-84ZN (phenazine non-producer, phzB::lacZ), 30-84PCA (PCA only producer, phzO::Tn5), 30-84O* (2OHPCA overproducer), 30-84H (PCA and PCN producer), 30-84M (PCA and 5MPCA producer), 30-84S (PCA and 1OHPZ producer), and 30-84MS (PCA and PYO producer). 30-84WT, 30-84ZN, 30-84PCA, and 30-84O* contain the empty vector pGT2-Ppsp.
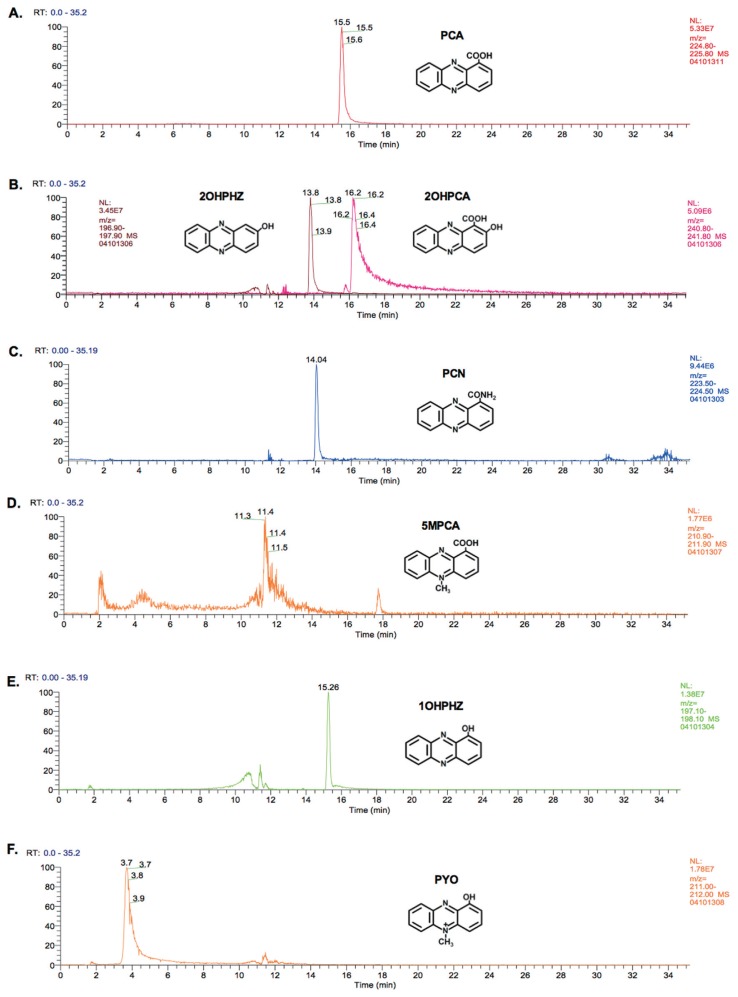
To further verify the production of different phenazines, total phenazines were extracted and separated by LC-MS according to monoisotopic mass, and the extracted ion chromatograms are shown (Fig. 2). One peak was detected in the phenazine extracts from 30-84PCA at RT = 15.5 min in m/z = 224–225 range, which corresponded to PCA (monoisotopic mass, 224.05 g/mol, respectively) (Fig. 2A). The identity of PCA was confirmed by comparison to extracts from strain P. synxantha 2-79 (formerly P. fluorescens), which produces only PCA (data not shown) (Mavrodi et al., 2001; Weller, 1983). The core PCA peak was detected in the extract of all the phenazine producing strains tested (data not shown). In addition to the PCA peak, phenazines extracted from 30-84WT demonstrated the characteristic 2 major peaks detected at RT = 13.8 min and 16.2 min in m/z = 196–197 and 240–241 ranges, respectively, corresponding to 2OHPHZ (196.06 g/mol) and 2OHPCA (240.05 g/mol) (Fig. 2B). Phenazines extracted from 30-84H contained a peak at RT = 14.1 min in m/z = 223–224 (Fig. 2C), corresponding to PCN (223.07 g/mol). The identity of PCN was confirmed by comparison to phenazines extracted from P. chlororaphis ATCC17411 (data not shown), previously shown to produce PCN (Mavrodi et al., 2001). Samples from 30-84M designed to produce 5MPCA had no clear peak other than for PCA, but instead numerous ambiguous “noisy peaks” were detected (Fig. 2D). This result is consistent with previous studies wherein 5MPCA could not be detected by C18-reverse phase HPLC extraction from either the phzS mutant of P. aeruginosa PAO1 or an E. coli strain expressing the phenazine biosynthetic operon and phzM in trans (Bellin et al., 2014; Mavrodi et al., 2001). Mavrodi et al. (2001) speculated that because 5MPCA is extremely hydrophilic, it is not effectively extracted and detected with the methods used. For this reason, production of 5MPCA of strain 30-84M was confirmed only by colorimetric observation as reported previously (Mavrodi et al., 2001). The phenazines extracted from 30-84S contained a peak corresponding to 1OHPHZ (196.06 g/mol) detected at RT = 15.53 in the range of m/z = 197–198 (Fig. 2E). Finally, in the extracts from 30-84MS, a peak corresponding to PYO (210.07 g/mol) was detected at RT = 3.7 min in the range of m/z = 211–212 (Fig. 2F). The identity of PYO was confirmed by comparison to a PYO standard (Sigma-Aldrich, St. Louis, MO, USA). Taken together, the results indicated that the introduced genes were converting PCA to the appropriate phenazine derivatives.
Fig. 2.
LC-MS analyses of phenazines extracted from isogenic derivatives of P. chlororaphis 30-84. (A) 30-84PCA, (B) 30-84WT, (C) 30-84H, (D) 30-84M, (E) 30-84S, and (F) 30-84MS. One microliter samples were used for the LC-MS separation and the identities of peaks were confirmed by monoisotopic mass analysis accompanied by comparisons to available positive control strains and standards (i.e., PCA only producer P. synxantha 2-79, PCN producer P. chlororaphis ATCC17411, and purified PYO). The assay was repeated three times.
Phenazines have specificity to inhibit the growth of plant pathogens
Phenazine production in P. chlororaphis 30-84 is responsible for the majority of its ability to inhibit the growth of the fungal pathogen Ggt (Pierson and Thomashow, 1992). To examine how the production of different phenazines affects the ability of P. chlororaphis 30-84 to inhibit plant pathogenic fungi, in vitro pathogen inhibition assays were performed. Only minor (less than 1.2 mm) or no inhibition was observed for the phenazine non-producing strain 30-84ZN (data not shown), whereas production of PCA only by 30-84PCA typically resulted in significantly more pathogen inhibition (Table 3). For all pathogens tested, the level of inhibition by 30-84PCA was used as the baseline for comparing the effectiveness of each additional phenazine derivative. Only 30-84WT significantly inhibited either Ggt or G. graminis var. graminis (Ggg), indicating the importance of 2OHPCA for inhibition of G graminis. Strain 30-84WT also had the broadest spectrum of pathogen inhibition. The production of different phenazines altered and broadened the spectrum of pathogens inhibited by P. chlororaphis 30-84 (Table 3). For example, 30-84MS most strongly inhibited the growth of Botrytis cinerea, Phytophthora capsici, and Sclerotinia sclerotiorum, whereas the PCN producing 30-84H and 1OHPZ producing 30-84S, most strongly inhibited the growth of Sclerotinia minor and Alternaria brassicae, respectively (Table 3). Given that the isogenic derivatives differed only in the type of phenazines produced, it appears that different phenazine derivatives differ in their abilities to inhibit the growth of different plant pathogenic fungi. Previous studies that compared the specificity of pathogen (bacterial, fungal, nematode) inhibition by strains producing different phenazines have similarly demonstrated differences in the spectrum of inhibitory targets (Chin-A-Woeng et al., 1998; Flaishman et al., 1990; Thomashow and Weller, 1988). However, the strains used in those studies also may differ from each other in other traits crucial to their interactions with specific pathogens. Our use of isogenic strains producing different phenazines enabled us to directly assess the spectrum of pathogen inhibition by various phenazine derivatives in a common background.
Table 3.
In vitro Fungal growth inhibition assays
| Pathogens | Inhibition zone (mm)a by altered phenazine producers (types of phenazine) | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| 30-84PCA | 30-84WT | 30-84H | 30-84M | 30-84S | 30-84MS | |
|
| ||||||
| (PCA) | (2OHPCA) | (PCN) | (5MPCA) | (1OHPZ) | (PYO) | |
| Alternaria brassicae | 6.5 ± 0.7 | 8.5 ± 1.41 | 6.5 ± 0.7 | 8 ± 1.41 | 9 ± 1.41* | 8 ± 1.41 |
| Botrytis cinerea | 4.5 ± 0.71 | 3.5 ± 0.71 | 2.5 ± 0.71 | 4.75 ± 0.35 | 3.5 ± 0.71 | 8.5 ± 2.12* |
| Cochliobolus heterotrophs | 1.25 ± 0.71 | 4.25 ± 0.75* | 1.25 ± 0.75 | 4.75 ± 0.85* | 3.75 ± 1.44* | 5.75 ± 0.75* |
| Colletotrichum graminicola | 6.25 ± 1.32 | 9.5 ± 0.95* | 7 ± 0.41 | 6.75 ± 0.75 | 6 ± 0.91 | 5.5 ± 0.5 |
| Fusrium graminearum | 1.3 ± 0.33 | 3.5 ± 0.65* | 2.15 ± 0.25* | 3 ± 0.57* | 1.38 ± 0.24 | 1.33 ± 0.33 |
| Fusarium oxysporum | 1.67 ± 0.28 | 3.5 ± 0.5* | 1.75 ± 0.25 | 2.25 ± 0.48 | 3.75 ± 0.28* | 2.25 ± 0.33 |
| Gaeumannomyces graminis var. graminis | 2.5 ± 0.4 | 8.5 ± 0.9* | 2 ± 0.4 | 3.4 ± 0.5 | 1.9 ± 0.3 | 2.8 ± 0.5 |
| Gaeumannomyces graminis var. tritici | 3.5 ± 0.3 | 7.5 ± 0.33* | 4.2 ± 0.36 | 3.1 ± 0.39 | 3.4 ± 0.29 | 3.3 ± 0.29 |
| Phytophthora capsici | 3.5 ± 0.71 | 3.45 ± 0.64 | 4.5 ± 0.71 | 2.5 ± 0.71 | 4 ± 0.14 | 6 ± 0.21* |
| Pythium ultimum | 1.5 ± 0.28 | 5.5 ± 0.14* | 2.4 ± 0.18* | 6.5 ± 0.5* | 3.15 ± 0.45* | 3.25 ± 0.63* |
| Rhizoctonia solani | 0 | 0.75 ± 0.47* | 0 | 1.25 ± 0.75* | 0 | 1 ± 0.58* |
| Sclerotinia minor | 4 ± 1.35 | 6.25 ± 1.31 | 7.25 ± 1.84* | 5.25 ± 1.11 | 2.75 ± 0.95 | 4.75 ± 1.31 |
| Sclerotinia sclerotiorum | 7.75 ± 1.18 | 8 ± 1.35 | 8 ± 1.35 | 8.75 ± 1.38 | 7.33 ± 1.86 | 11.25 ± 0.75* |
Mean width of the zones of inhibition (mm) obtained from mean of 3 separate assays ± standard error, n = 9.
Asterisk (*) with gray highlighted cells indicate values that differ significantly from 30-84PCA.
The mechanisms for the antimicrobial activities of phenazines have not been fully elucidated but appear to be related to their capacity to generate reactive oxygen species (ROS) and are likely affected by the chemical properties of each specific phenazine derivative, influencing their redox activity and solubility under different pH conditions (Cezairliyan et al., 2013; Gibson et al., 2009; Hassett et al., 1992; Liu and Nizet, 2009; Morales et al., 2010; Ownley et al., 1992; Pierson and Pierson, 2010; Price-Whelan et al., 2006). For example, 5MPCA produced by P. aeruginosa PA14 showed stronger antifungal activity against Candida albicans than PCA or PYO (Gibson et al., 2009; Morales et al., 2010). The authors reported that 5MPCA was covalently modified by amino acids in the fungal cytoplasm and accumulated extensively, resulting in the generation of ROS within fungal cells (Gibson et al., 2009). A role for pH in in vitro antifungal activity has been demonstrated in several studies. For example, growth inhibition of Ggt due to purified PCA was greater at pH 6.0 than at higher pH values (Ownley et al., 1992), whereas inhibition of F. oxysporum f. sp. radicis-lycopersici due to purified PCN was 10 times greater than PCA at pH 5.7 (Chin-A-Woeng et al., 1998). The ability of PCA or PYO produced by P. aeruginosa PA14 to kill the nematode Caenorhabditis elegans was highly dependent on pH (Cezairliyan et al., 2013). The authors speculated that antibiotic activity might be influenced by the acid dissociation constants (pKa) of each phenazine derivative affecting their ionic form or solubility (Cezairliyan et al., 2013; Chin-A-Woeng et al., 1998).
Phenazines and their role in take-all disease suppression
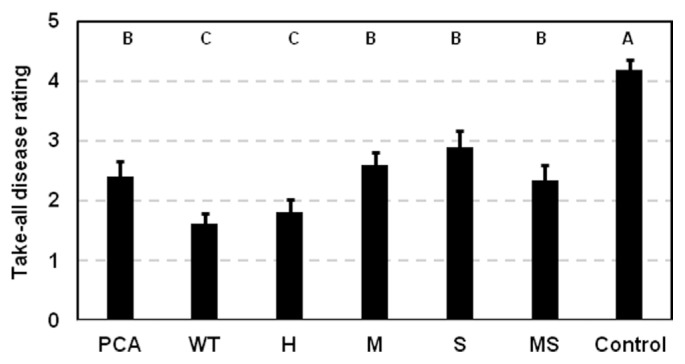
To determine whether modifying the type of phenazines produced altered take-all disease suppression, take-all disease symptoms on plants inoculated with the altered phenazine-producing strains were compared to plants inoculated with 30-84WT, 30-84PCA, or a control receiving no bacteria. The strains were introduced as a methyl cellulose inoculum onto seeds that were then grown in autoclaved soil subsequently infested with Ggt. There were no statistical differences among the strains in their ability to colonize and persist on roots for twenty days: all reached populations of 107 cfu/gram of root dry weight (data not shown). As expected, 30-84WT inoculated roots had significantly reduced take-all symptoms compared to 30-84PCA (Fig. 3). Roots inoculated with most of the altered phenazine producers displayed disease symptoms similar to those observed on plants inoculated with 30-84PCA. The exception was 30-84H (PCN producer), which reduced take-all symptoms on roots similar to 30-84WT, despite not being as effective as 30-84WT in inhibiting Ggt in vitro (Table 3).
Fig. 3.
Suppression of take-all disease symptoms on wheat roots by isogenic derivatives of P. chlororaphis 30-84. 30-84WT, 30-84PCA and altered phenazine produces were seed inoculated and planted into a soil mix infested with Ggt. After 20 days of growth, roots were thoroughly washed and evaluated for disease severity on scale of 0 (no disease) – 5 (nearly dead). The values are the mean of two separate experiments (8 plants per each experiment) with standard error bars. Values with the same letter do not differ significantly as determined by a Fishers protected Least Significantly Difference (LSD) test (P > 0.05).
Phenazines and their roles in biofilm formation
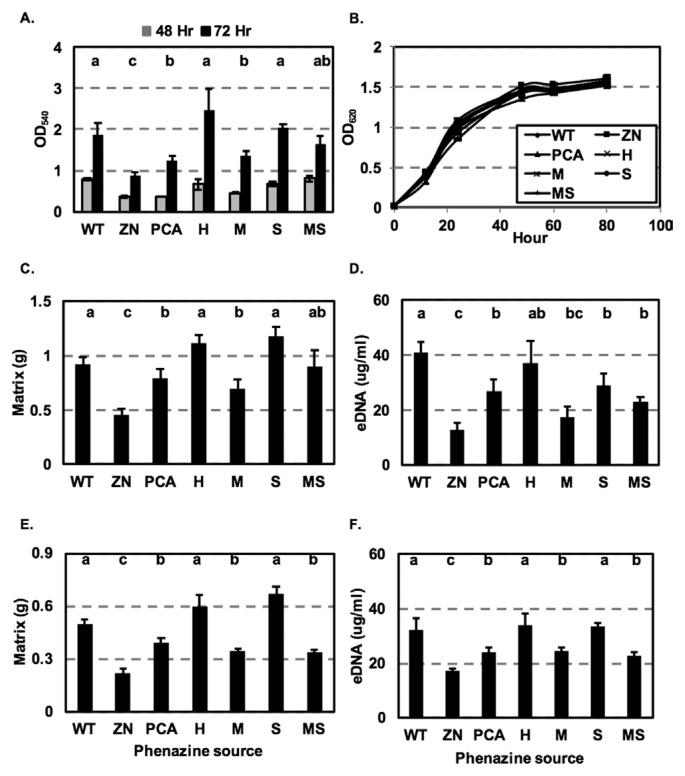
The essential role of phenazines in biofilm formation was demonstrated in studies in which phenazine-defective mutants were significantly impaired in cell attachment and biofilm formation as compared to wild type. For example, Maddula et al. (2006) showed that the phenazine non-producing strain 30-84ZN was significantly impaired in biofilm formation compared to 30-84WT. Similarly, the importance of phenazine for biofilm production and morphology was shown using P. aeruginosa PA14 and a phenazine-defective mutant (Ramos et al., 2010). We hypothesized that the production of different phenazines by the isogenic derivatives may alter biofilm formation by P. chlororaphis 30-84. To test this hypothesis, 30-84WT, 30-84ZN and the altered phenazine-producing strains were grown without shaking in 96-well polystyrene plates and the populations of cells attached to the well surfaces were determined. The phenazine non-producing strain 30-84ZN was significantly impaired in biofilm formation after 48 h compared to the phenazine producers, whereas all of the phenazine-producing strains formed similar amounts of biofilm. After 72 h, biofilm formation by 30-84ZN remained impaired compared to the phenazine producers. Notably strains 30-84WT, 30-84H, and 30-84S produced significantly more biofilm than 30-84PCA and 30-84M, whereas the amount of biofilm produced by 30-84MS was not statistically different from any of the other phenazine-producing strains (Fig. 4A). However, there were no differences between 30-84WT, 30-84ZN and the altered phenazine producing strains in their growth rates (Fig. 4B), suggesting that the variation in the amount of biofilm formed by these strains was not a result of alterations in bacterial growth. The results indicated that some of the different phenazines produced by strain 30-84 affected biofilm formation. It is unknown whether the hydrophilicity difference between 5MPCA and the other derivatives may have contributed to the lower amount of biofilm formed by 30-84M.
Fig. 4.
Growth curve, biofilm formation and eDNA production by isogenic derivatives of P. chlororaphis 30-84. (A) Biofilm production by cultures grown in 96-well plates containing AB-C for 48 and 72 h. Attached cells with crystal violet staining were quantified at optical density 540 nm. (B) Bacterial cultures were grown in 96-well microtiter polystyrene plates containing AB-C up to 80 h without agitation. Cell density was measured periodically by optical density of 620 nm. (C) Quantification of non-attached floating biofilm matrix by cultures grown in 24-well plates containing AB-C for 72 h at 28°C without agitation. Non-attached floating biofilm matrix was determined by weight. (D) Quantification of extracellular DNA produced by cultures grown in 24-well polystyrene plates containing AB-C for 72 h at 28°C without agitation; extracellular DNA concentration was quantified using a Qubit fluorometer (invitrogen Life Technologies). Quantification of (E) non-attached floating biofilm matrix and (F) extracellular DNA produced by the phenazine non-producing strain 30-84ZN when grown for 72 h in cultures containing supernatants from each of the phenazine-producing strains or 30-84ZN (negative control). The designation on the X axis indicates the source of the supernatants. All data represent the average of 6 biological replicates and error bars indicate the standard error. Values with the same letter do not differ significantly as determined by a Fishers protected Least Significantly Difference (LSD) test (P > 0.05).
In addition to forming surface attached populations, after 72 h substantial numbers of cells also survived in non-attached floating biofilms as described previously for P. aeruginosa (Alhede et al., 2011). Previous work using P. chlororaphis 30-84 showed that 2OHPCA greatly enhanced the production of structured matrix in floating biofilms and that this was related to extracellular DNA (eDNA) release (Wang et al., 2016). In the present study, 30-84WT, 30-84ZN, and the altered phenazine-producing strains were grown without shaking in 24-well plates and the non-attached floating biofilm matrix was quantified. The amount of biofilm matrix produced by 30-84ZN was significantly lower than the amount of produced by all phenazine producing strains after 72 h (Fig. 4C). Similarly, the amount of eDNA produced by 30-84ZN (12.63 ± 2.7 g/ml) after 72 h was significantly lower than the amount produced by all of the phenazine-producing strains, confirming the important role of phenazine in eDNA release (Fig. 4D). Production of eDNA was generally correlated with the amount of biofilm produced, being highest for 30-84WT (41.07 ± 3.6 μg/ml) and 30-84H (37.3 ± 7.7 μg/ml), less for 30-84S (28.82 ± 4.3 μg/ml), 30-84PCA (26.96 ± 4.0 μg/ml), and 30-84MS (23.1 ± 1.2 μg/ml) and significantly less for 30-84M (17.6 ± 5.8 μg/ml). The results indicate that the production of different phenazines affects the amount of eDNA released in a manner that is correlated with biofilm matrix production.
To verify the role of different phenazines in biofilm matrix and eDNA production, 30-84ZN was inoculated into medium amended with cell-free, DNA-free (treated with DNase I), phenazine-containing supernatants from either 30-84WT, or the altered phenazine-producing strains. Biofilm matrix production by 30-84ZN grown in medium with 30-84ZN supernatant (control treatment) was significantly impaired, whereas biofilm matrix production by 30-84ZN grown in medium containing supernatants of 30-84WT, 30-84H or 30-84S was significantly greater than when grown in supernatants of 30-84PCA, 30-84M or 30-84MS (Fig. 4E). Addition of 30-84ZN supernatant to the medium resulted in the production of eDNA (17.13 ± 1.03 μg/ml) at a level similar to that produced by 30-84ZN in the absence of supernatant (Fig. 4D, 4F). However, growth of 30-84ZN in the phenazine-containing supernatants significantly increased the amount eDNA produced in a manner generally correlated with the amount of biofilm produced by the strains comprising each supernatant. The amounts of eDNA produced by 30-84ZN grown in medium containing supernatants of 30-84WT (32.2 ± 4.4 μg/ml), 30-84H (34.03 ± 4.1 μg/ml) or 30-84S (33.73 ± 1.5 μg/ml) were significantly higher than when 30-84ZN was grown in medium containing supernatants of 30-84PCA (24.1 ± 1.6 μg/ml), 30-84M (24.55 ± 1.48 μg/ml) or 30-84MS (22.78 ± 1.5 μg/ml) (Fig. 4F). Although it cannot be concluded that the observed results using cell free, DNA-free supernatants are due solely to the different type of phenazine produced by the isogenic derivatives, the results are consistent with the observation that different phenazines consistently promote the production of different amounts of eDNA and biofilm matrix.
A role for eDNA in the establishment of bacterial biofilms has been recognized for nearly a decade, however the source of the DNA remains controversial. One hypothesis is that eDNA in the biofilm matrix is derived from chromosomal DNA resulting from cell lysis, which may be mediated by the generation of ROS (Okshevsky and Meyer, 2015; Wang et al., 2016; Wei and Ma, 2013). Das and Manefield (2012) suggested that phenazines may lead to the generation and accumulation of ROS, which in turn may cause damage to the bacterial cell membrane, cell lysis, and release of chromosomal DNA into the extracellular space as eDNA. Recently, it was demonstrated that release of eDNA by P. chlororaphis 30-84 and 30-84O* (overproducing 2OHPCA) was greater than by 30-84PCA or 30-84ZN (Wang et al., 2016). Moreover, whole-transcriptomic analysis showed that the expression of genes encoding ROS detoxifying enzymes and DNA repair/modification enzymes were significantly higher for phenazine producing strains (especially 2OHPCA producers) than for the phenazine deficient strain. In that study, it was postulated that eDNA release may be related in part to the activity of a chromosomally encoded bacterial pyocin, the production of which increased with ROS generation (Wang et al., 2016). Similarly, studies using wild type P. aeruginosa and phenazine-deficient mutants demonstrated that phenazine production (i.e., PYO) contributed to eDNA release and was correlated with the generation of H2O2 and cell lysis (Das et al., 2013a; Das and Manefield, 2012). Since different phenazines have different redox potentials (Bellin et al., 2014; Price-Whelan et al., 2006; Wang and Newman, 2008), it is reasonable to speculate that the efficiency of cell lysis and subsequent eDNA release might be dependent on what types of phenazine the cells produce and their effect on ROS generation.
Role of phenazines in colonization as rhizosphere persistence
To determine whether specific phenazines contribute to root colonization and rhizosphere persistence, 30-84WT, 30-84ZN, 30-84PCA, and the altered phenazine-producing strains were inoculated separately into natural wheat field soil (containing the native soil microbiota) and populations colonizing the roots from the soil inoculant were determined after repeated plant/harvest cycles. The bacterial populations of 30-84ZN on roots decreased by one log unit over five repeat harvests whereas the bacterial populations of the phenazine producers remained unchanged (Table 4). Although this was not a dramatic decline, our findings are in agreement with previous findings in which the population of a phenazine deficient mutant declined more rapidly than wild-type over repeated plant/harvest cycles in natural soil (Mazzola et al., 1992). These results indicate that although phenazine production is important for bacterial colonization of roots and persistence in the rhizosphere over repeated harvest/colonization cycles, the type of phenazines produced is less important than whether cells produce phenazines under these conditions.
Table 4.
Role of altered phenazine production on root colonization and persistence
| Plant/harvest cycle | Bacterial Population (Log cfu/g of root dry weight)a | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| WT | ZN | PCA | H | M | S | MS | |
| 1 | 7.7 ± 0.1A | 7.3 ± 0.3A | 7.6 ± 0.2A | 7.6 ± 0.2A | 7.5 ± 0.2A | 7.4 ± 0.1A | 7.2 ± 0.3A |
| 2 | 7.3 ± 0.1A | 7.5 ± 0.2A | 7.2 ± 0.1A | 7.2 ± 0.3A | 7.1 ± 0.4A | 7.0 ± 0.3A | 7.0 ± 0.3A |
| 3 | 7.5 ± 0.2A | 7.4 ± 0.2A | 7.4 ± 0.1A | 7.6 ± 0.1A | 7.2 ± 0.1A | 7.4 ± 0.1A | 7.5 ± 0.1A |
| 4 | 7.4 ± 0.2A | 6.7 ± 0.1B | 7.2 ± 0.1A | 7.1 ± 0.1A | 7.1 ± 0.2A | 7.3 ± 0.2A | 7.4 ± 0.2A |
| 5 | 7.2 ± 0.2A | 6.1 ± 0.2B | 7.1 ± 0.2A | 7.1 ± 0.2A | 7.0 ± 0.3A | 7.1 ± 0.2A | 6.9 ± 0.2A |
Pregerminated wheat seedlings were planted in natural soil mix inoculated with 30-84WT, 30-84ZN, and altered phenazine-producing strains. Initial bacterial population of each strain was approximately 108 cfu/g of soil, and population was determined after each 20-day cycle. Data represent the means of 6 separate plants ± standard errors. The values with the different letters indicate statistically significant differences in each cycle as determined by Fishers protected Least Significantly Difference (LSD) tests (P < 0.05).
Indigenous phenazine-producing pseudomonads are abundant in agricultural soils (Mavrodi et al., 2010). In the intermountain region of the northwestern United States, it was reported that the frequency of wheat root systems colonized by phenazine-producing (Phz+) pseudomonads was inversely related to annual precipitation (Mavrodi et al., 2012a, 2012b). Phz+ pseudomonads were shown to be abundant in the low-precipitation zone and on wheat roots from dryland production, with populations ranging from 5 to 6.37 log cfu/g root and rhizosphere colonization frequencies of 67 to 100 percent (Mavrodi et al., 2012a, 2012b). Moreover, the populations of indigenous Phz+ wheat-colonizing pseudomonads were genotypically and phenotypically diverse (Parejko et al., 2012). In these studies, the abundance of phenazine producers was based on the prevalence of the phzF gene, which is not a precise predictor of the ability to produce phenazines or the type of phenazines produced (Parejko et al., 2012). It is logical to assume that this indigenous population would include strains that produce PCA as well as strains capable of producing one or more additional types of phenazines. This hypothesis is supported by our observations that when introduced into soil, rhizosphere colonization and persistence were diminished only in strains without the capacity to produce phenazines (e.g., 30-84ZN) and not by the type of phenazines they produced. However, our data suggest that not all phenazines produced by this indigenous Phz+ population are equally effective in their ability to suppress root take-all disease due to Ggt infection. We showed that production of the 2-hydroxy derivatives and PCN resulted in the most effective take-all disease suppression under our conditions. With regard to biological control applications, we speculate that phenazine-producers introduced for biological control are likely to establish well in low moisture production zones. There introduced and indigenous strains may form a combined community supportive of all phenazine production via the exchange of quorums sensing signals (Pierson et al., 1998) and in which strains that produce phenazines important for take-all suppression need only be a stable component. Thus, future strategies for improving the use of phenazine-producing strains for biological control may need to consider both the environment in which these strains are likely to thrive as well agricultural practices that encourage sustainable indigenous phenazine-producing populations.
Acknowledgments
We thank Dr. Dimitri Mavrodi (University of Southern Mississippi) and Dr. Linda Thomashow (USDA-ARS Root Disease and Biological Control Research Center) for generously providing information and helpful discussions regarding the cloning of the phenazine modifying genes and phenazine purification. We thank Dr. Yohannes Rezenom at Laboratory for biological mass spectrometry, Texas A&M University for the assistance of the purification and identification of phenazines. We thank Dr. Dave Weller (USDA-ARS Root Disease and Biological Control Research Center) for generously contributing virulent Ggt. This project was supported in part by United States Department of Agriculture (USDA) National Institute of Food and Agriculture (NIFA) award no. 2008-35319-04490 and other funding to E. Pierson.
References
- Alhede M, Kragh KN, Qvortrup K, Allesen-Holm M, van Gennip M, Christensen LD, Jensen PØ, Nielsen AK, Parsek M, Wozniak D. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS One. 2011;6:e27943. doi: 10.1371/journal.pone.0027943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron SS, Terranova G, Rowe JJ. Molecular mechanism of the antimicrobial action of pyocyanin. Curr Microbiol. 1989;18:223–230. doi: 10.1007/BF01570296. [DOI] [Google Scholar]
- Bellin DL, Sakhtah H, Rosenstein JK, Levine PM, Thimot J, Emmett K, Dietrich LE, Shepard KL. Integrated circuit-based electrochemical sensor for spatially resolved detection of redox-active metabolites in biofilms. Nat Commun. 2014;5:3256. doi: 10.1038/ncomms4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G, Fritze A, Roskot N, Smalla K. Evaluation of potential biocontrol rhizobacteria from different host plants of Verticillium dahliae kleb. J Appl Microbiol. 2001;91:963–971. doi: 10.1046/j.1365-2672.2001.01462.x. [DOI] [PubMed] [Google Scholar]
- Cezairliyan B, Vinayavekhin N, Grenfell-Lee D, Yuen GJ, Saghatelian A, Ausubel FM. Identification of Pseudomonas aeruginosa phenazines that kill Caenorhabditis elegans. PLoS Pathog. 2013;9:e1003101. doi: 10.1371/journal.ppat.1003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin-A-Woeng TF, Bloemberg GV, Lugtenberg BJ. Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol. 2003;157:503–523. doi: 10.1046/j.1469-8137.2003.00686.x. [DOI] [PubMed] [Google Scholar]
- Chin-A-Woeng TF, Bloemberg GV, van der Bij AJ, van der Drift KM, Schripsema J, Kroon B, Scheffer RJ, Keel C, Bakker PA, Tichy H-V. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis pcl1391 of tomato root rot caused by Fusarium oxysporum f. Sp. radicis-lycopersici. Mol Plant-Microbe Interact. 1998;11:1069–1077. doi: 10.1094/MPMI.1998.11.11.1069. [DOI] [Google Scholar]
- Chin-A-Woeng TF, Thomas-Oates JE, Lugtenberg BJ, Bloemberg GV. Introduction of the phzh gene of Pseudomonas chlororaphis pcl1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol Plant-Microbe Interact. 2001a;14:1006–1015. doi: 10.1094/MPMI.2001.14.8.1006. [DOI] [PubMed] [Google Scholar]
- Chin-A-Woeng TF, van den Broek D, de Voer G, van der Drift KM, Tuinman S, Thomas-Oates JE, Lugtenberg BJ, Bloemberg GV. Phenazine-1-carboxamide production in the biocontrol strain Pseudomonas chlororaphis pcl1391 is regulated by multiple factors secreted into the growth medium. Mol Plant-Microbe Interact. 2001b;14:969–979. doi: 10.1094/MPMI.2001.14.8.969. [DOI] [PubMed] [Google Scholar]
- Das T, Kutty SK, Kumar N, Manefield M. Pyocyanin facilitates extracellular DNA binding to Pseudomonas aeruginosa influencing cell surface properties and aggregation. PLoS One. 2013a;8:e58299. doi: 10.1371/journal.pone.0058299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Kutty SK, Tavallaie R, Ibugo AI, Panchompoo J, Sehar S, Aldous L, Yeung AW, Thomas SR, Kumar N. Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm formation. Sci Rep. 2015;5:8398. doi: 10.1038/srep08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Manefield M. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS One. 2012;7:e46718. doi: 10.1371/journal.pone.0046718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Sehar S, Manefield M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ Microbiol Rep. 2013b;5:778–786. doi: 10.1111/1758-2229.12085. [DOI] [PubMed] [Google Scholar]
- Das T, Sharma PK, Busscher HJ, van der Mei HC, Krom BP. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl Environ Microbiol. 2010;76:3405–3408. doi: 10.1128/AEM.03119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney SM, Mavrodi DV, Bonsall RF, Thomashow LS. Phzo, a gene for biosynthesis of 2-hydroxylated phenazine compounds in Pseudomonas aureofaciens 30–84. J Bacteriol. 2001;183:318–327. doi: 10.1128/JB.183.1.318-327.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaishman M, Eyal Z, Voisard C, Haas D. Suppression of Septoria tritici by phenazine-or siderophore-deficient mutants of Pseudomonas. Curr Microbiol. 1990;20:121–124. doi: 10.1007/BF02092884. [DOI] [Google Scholar]
- Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Ghosh PK, Maiti TK. Structure of extracellular polysaccharides (eps) produced by rhizobia and their functions in legume-bacteria symbiosis. Achiev Life Sci. 2016;10:136–143. doi: 10.1016/j.als.2016.11.003. [DOI] [Google Scholar]
- Gibson J, Sood A, Hogan DA. Pseudomonas aeruginosa-candida albicans interactions: localization and fungal toxicity of a phenazine derivative. Appl Environ Microbiol. 2009;75:504–513. doi: 10.1128/AEM.01037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, Mililli L, Hunt C, Lu J, Osvath SR. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci USA. 2013;110:11541–11546. doi: 10.1073/pnas.1218898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Imlay JA. The soxrs response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol Microbiol. 2011;79:1136–1150. doi: 10.1111/j.1365-2958.2010.07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn JS, Bakaletz LO, Wozniak DJ. What’s on the outside matters: the role of the extracellular polymeric substance of gram-negative biofilms in evading host immunity and as a target for therapeutic intervention. J Biol Chem. 2016;291:12538–12546. doi: 10.1074/jbc.R115.707547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D, Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- Hassett D, Charniga L, Bean K, Ohman D, Cohen MS. Response of Pseudomonas aeruginosa to pyocyanin: mechanisms of resistance, antioxidant defenses, and demonstration of a manganese-cofactored superoxide dismutase. Infect Immun. 1992;60:328–336. doi: 10.1128/iai.60.2.328-336.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes WC, Stodola FH, Locke JM, Pridham TG, Conway HF, Sohns VE, Jackson RW. Pseudomonas aureofaciens kluyver and phenazine α-carboxylic acid, its characteristic pigment. J Bacteriol. 1956;72:412. doi: 10.1128/jb.72.3.412-417.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayathilake PG, Jana S, Rushton S, Swailes D, Bridgens B, Curtis T, Chen J. Extracellular polymeric substance production and aggregated bacteria colonization influence the competition of microbes in biofilms. Front Microbiol. 2017;1865;8 doi: 10.3389/fmicb.2017.01865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GY, Nizet V. Color me bad: microbial pigments as virulence factors. Trends Microbiol. 2009;17:406–413. doi: 10.1016/j.tim.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddula VS, Pierson EA, Pierson LS., III Altering the ratio of phenazines in Pseudomonas chlororaphis (aureofaciens) strain 30-84: effects on biofilm formation and pathogen inhibition. J Bacteriol. 2008;190:2759–2766. doi: 10.1128/JB.01587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddula VS, Zhang Z, Pierson EA, Pierson LS., III Quorum sensing and phenazines are involved in biofilm formation by Pseudomonas chlororaphis (aureofaciens) strain 30-84. Microb Ecol. 2006;52:289–301. doi: 10.1007/s00248-006-9064-6. [DOI] [PubMed] [Google Scholar]
- Mann EE, Wozniak DJ. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev. 2012;36:893–916. doi: 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi DV, Blankenfeldt W, Thomashow LS. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol. 2006;44:417–445. doi: 10.1146/annurev.phyto.44.013106.145710. [DOI] [PubMed] [Google Scholar]
- Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa pao1. J Bacteriol. 2001;183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi DV, Mavrodi OV, Parejko JA, Bonsall RF, Kwak Y-S, Paulitz TC, Thomashow LS, Weller DM. Accumulation of the antibiotic phenazine-1-carboxylic acid in the rhizosphere of dryland cereals. Appl Environ Microbiol. 2012a;78:804–812. doi: 10.1128/AEM.06784-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi DV, Peever TL, Mavrodi OV, Parejko JA, Raaijmakers JM, Lemanceau P, Mazurier S, Heide L, Blankenfeldt W, Weller DM. Diversity and evolution of the phenazine biosynthesis pathway. Appl Environ Microbiol. 2010;76:866–879. doi: 10.1128/AEM.02009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi OV, Mavrodi DV, Parejko JA, Thomashow LS, Weller DM. Irrigation differentially impacts populations of indigenous antibiotic-producing Pseudomonas spp. in the rhizosphere of wheat. Appl Environ Microbiol. 2012b;78:3214–3220. doi: 10.1128/AEM.07968-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzola M, Cook RJ, Thomashow LS, Weller DM, Pierson LS., III Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl Environ Microbiol. 1992;58:2616–2624. doi: 10.1128/aem.58.8.2616-2624.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WG, Leveau JH, Lindow SE. Improved gfp and inaz broad-host-range promoter-probe vectors. Mol Plant-Microbe Interact. 2000;13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- Morales DK, Jacobs NJ, Rajamani S, Krishnamurthy M, Cubillos-Ruiz JR, Hogan DA. Antifungal mechanisms by which a novel Pseudomonas aeruginosa phenazine toxin kills candida albicans in biofilms. Mol Microbiol. 2010;78:1379–1392. doi: 10.1111/j.1365-2958.2010.07414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens wcs365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Okshevsky M, Meyer RL. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol. 2015;41:341–352. doi: 10.3109/1040841X.2013.841639. [DOI] [PubMed] [Google Scholar]
- Ownley BH, Weller D, Thomashow LS. Influence of in situ and in vitro ph on suppression of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens 2-79. Phytopathology. 1992;82:178–184. doi: 10.1094/Phyto-82-178. [DOI] [Google Scholar]
- Parejko JA, Mavrodi DV, Mavrodi OV, Weller DM, Thomashow LS. Population structure and diversity of phenazine-1-carboxylic acid producing fluorescent Pseudomonas spp. from dryland cereal fields of central washington state (USA) Microb Ecol. 2012;64:226–241. doi: 10.1007/s00248-012-0015-0. [DOI] [PubMed] [Google Scholar]
- Pierson EA, Wood DW, Cannon JA, Blachere FM, Pierson LS., III Interpopulation signaling via n-acyl-homoserine lactones among bacteria in the wheat rhizosphere. Mol Plant-Microbe Interact. 1998;11:1078–1084. doi: 10.1094/MPMI.1998.11.11.1078. [DOI] [Google Scholar]
- Pierson LS, III, Gaffney T, Lam S, Gong F. Molecular analysis of genes encoding phenazine biosynthesis in the biological control bacterium pseudomonas aureofaciens 30-84. FEMS Microbiol Lett. 1995;134:299–307. doi: 10.1111/j.1574-6968.1995.tb07954.x. [DOI] [PubMed] [Google Scholar]
- Pierson LS, III, Pierson EA. Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol. 2010;86:1659–1670. doi: 10.1007/s00253-010-2509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson LS, III, Thomashow LS. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30-84. Mol Plant-Microbe Interact. 1992;5:330–339. doi: 10.1094/MPMI-5-330. [DOI] [PubMed] [Google Scholar]
- Price-Whelan A, Dietrich LE, Newman DK. Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- Ramos I, Dietrich LE, Price-Whelan A, Newman DK. Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res Microbiol. 2010;161:187–191. doi: 10.1016/j.resmic.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; NY, USA: 2001. [Google Scholar]
- Selin C, Habibian R, Poritsanos N, Athukorala SN, Fernando D, De Kievit TR. Phenazines are not essential for Pseudomonas chlororaphis pa23 biocontrol of Sclerotinia sclerotiorum, but do play a role in biofilm formation. FEMS Microbiol Ecol. 2009;71:73–83. doi: 10.1111/j.1574-6941.2009.00792.x. [DOI] [PubMed] [Google Scholar]
- Steinberg N, Kolodkin-Gal I. The matrix reloaded: How sensing the extracellular matrix synchronizes bacterial communities. J Bacteriol. 2015;197:2092–2103. doi: 10.1128/JB.02516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow LS, Weller DM. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J Bacteriol. 1988;170:3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM, Messenger AJ. Occurrence, biochemistry and physiology of phenazine pigment production. Adv Microb Physiol. 1986;27:211–275. doi: 10.1016/S0065-2911(08)60306-9. [DOI] [PubMed] [Google Scholar]
- Wang D, Yu JM, Dorosky RJ, Pierson LS, III, Pierson EA. The phenazine 2-hydroxy-phenazine-1-carboxylic acid promotes extracellular DNA release and has broad transcriptomic consequences in Pseudomonas chlororaphis 30-84. PLoS One. 2016;11:e0148003. doi: 10.1371/journal.pone.0148003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Newman DK. Redox reactions of phenazine antibiotics with ferric (hydr) oxides and molecular oxygen. Envrion Sci Technol. 2008;42:2380–2386. doi: 10.1021/es702290a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol. 2011;193:3606–3617. doi: 10.1128/JB.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Ma LZ. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int J Mol Sci. 2013;14:20983–21005. doi: 10.3390/ijms141020983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller D. Colonization of wheat roots by a fluorescent pseudomonad suppressive to take-all. Phytopathology. 1983;73:1548–1553. doi: 10.1094/Phyto-73-1548. [DOI] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487–1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Wilkinson H, Cook R, Alldredge J. Relation of inoculum size and concentration to infection of wheat roots by Gaeumannomyces graminis var. tritici. Phytopathology. 1985;75:98–103. doi: 10.1094/Phyto-75-98. [DOI] [Google Scholar]
- Wood DW, Gong F, Daykin MM, Williams P, Pierson LS., III N-acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. J Bacteriol. 1997;179:7663–7670. doi: 10.1128/jb.179.24.7663-7670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JM. PhD thesis. Texas A&M University; College Station, TX, USA: 2016. Regulation and ecological roles of phenazine biosynthesis in the biological control strain Pseudomonas chlororaphis 30-84. [Google Scholar]
- Yu JM, Wang D, Pierson LS, III, Pierson EA. Disruption of MiaA provides insights into the regulation of phenazine biosynthesis under suboptimal growth conditions in Pseudomonas chlororaphis 30-84. Microbiology. 2017;163:94–108. doi: 10.1099/mic.0.000409. [DOI] [PubMed] [Google Scholar]
- Zhou L, Jiang H-X, Sun S, Yang D-D, Jin K-M, Zhang W, He Y-W. Biotechnological potential of a rhizosphere Pseudomonas aeruginosa strain producing phenazine-1-carboxylic acid and phenazine-1-carboxamide. World J Micriobiol Biotech. 2016;32:50. doi: 10.1007/s11274-015-1987-y. [DOI] [PubMed] [Google Scholar]