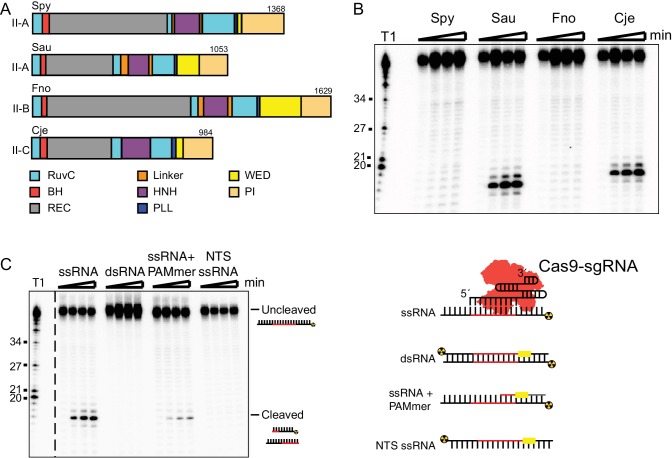
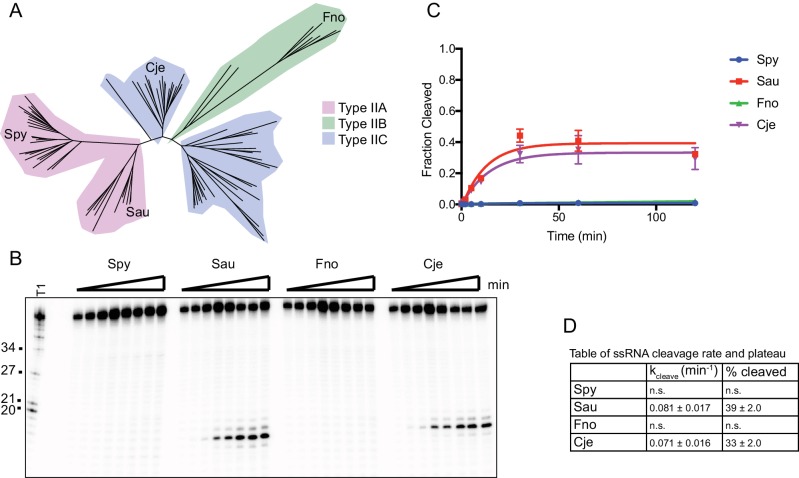
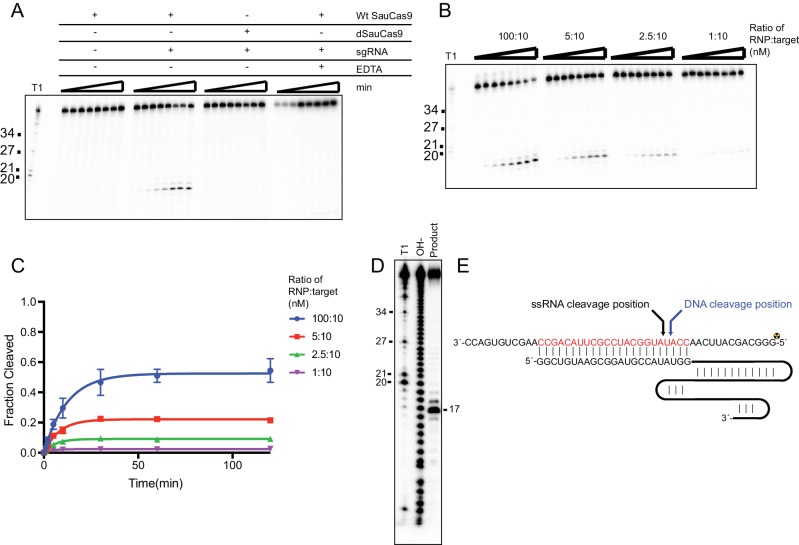
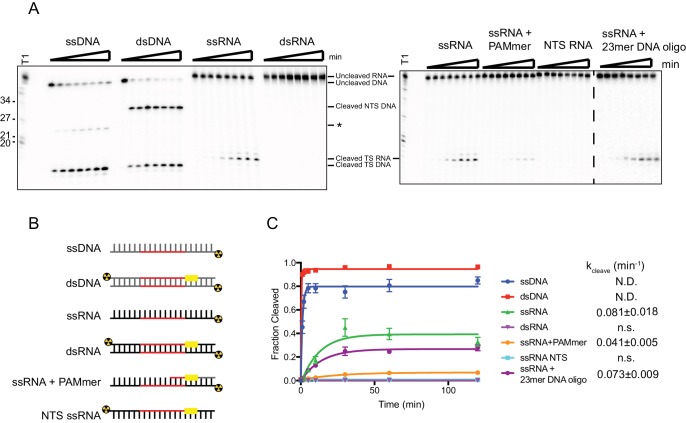
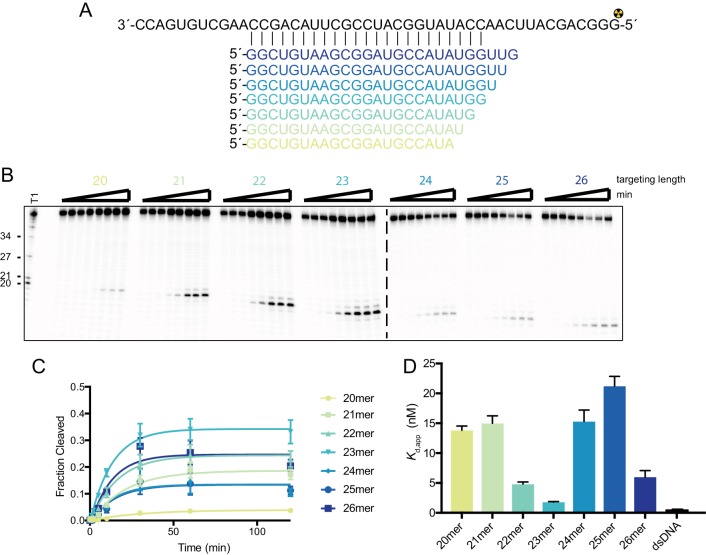
Figure 1. SauCas9 cleaves single-stranded RNA without a PAMmer.
(A) Schematic of Cas9 proteins tested for sgRNA-mediated RNA cleavage. RuvC, RuvC nuclease domain; BH, bridge-helix; REC, recognition domain; HNH, HNH nuclease domain; PLL, phosphate-lock loop; WED, wedge domain; PI, PAM-interacting domain. Adapted from (Nishimasu et al., 2014; 2015; Hirano et al., 2016; Yamada et al., 2017). (B) Representative in vitro cleavage of ssRNA by Cas9-sgRNA RNP complexes of homologs in (A). Radiolabeled pUC target RNA was incubated with Cas9 RNP at 37˚C and time points were taken at 0, 10, 30, and 60 min. Full time course is presented in Figure 1—figure supplement 1B. T1 indicates size markers generated by RNase T1 digestion of ssRNA target. Size in nucleotides is indicated on the left. (C) (Right) In vitro cleavage assay of various RNA substrates (Left). Full time course is presented in Figure 1—figure supplement 3A.