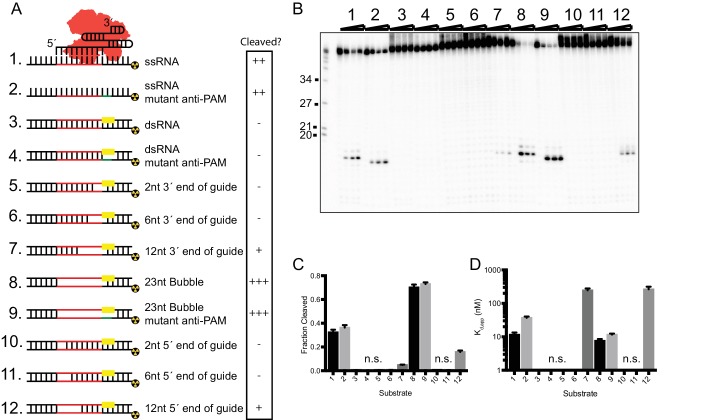
Figure 2. In vitro RNA cleavage is impaired by strong secondary structure.
(A) Schematic representation of structured RNA targets for in vitro cleavage assays. Symbols on right indicate relative level of cleavage activity for each substrate: ‘-”, no cleavage; ‘+”, low cleavage; ‘++”, medium cleavage; ‘+++” high cleavage. (B) Representative cleavage assay of partially-duplexed RNA targets diagrammed in (A). T1 indicates size markers generated by RNase T1 digestion of ssRNA target. Size in nucleotides is indicated on the left. (C and D) Fraction of target cleaved (C) and Kd,app (D) for substrates diagrammed in (A). Fits were determined in Prism using a single-exponential decay and a one-site binding model, respectively. Bars represent mean ± S.D. (n = 3). N.s. denotes no significant cleavage or binding.