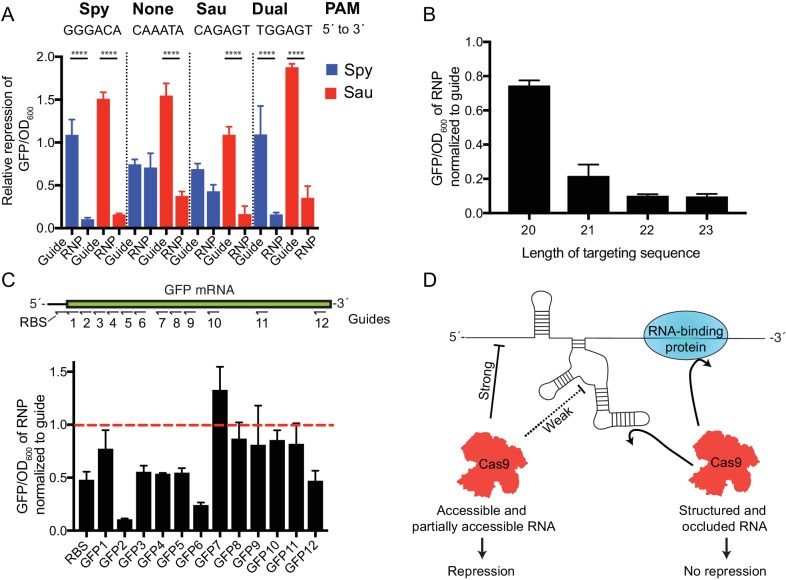
Figure 4. SauCas9 repression of a GFP reporter in vivo.
(A) Comparison of dSpy and dSauCas9 to repress GFP expression on the DNA and RNA level. GFP signal is normalized to OD600 to control for difference in cell density between samples. GFP/OD600 ratios for guide alone and RNP are normalized to values for a non-targeting guide vector and an Apo protein control, respectively. Target sites were chosen to be adjacent to PAM sites for Spy, Sau, both, or neither as indicated. Note: the slight GFP repression observed with dSpyCas9 using the target sequence adjacent to the Sau PAM (CAGAGT) likely results from the ability of SpyCas9 to use an NAG PAM, albeit with reduced efficiency (Hsu et al., 2013). ****p<0.0001 by one-way ANOVA. (B) Relative expression of GFP using guides with different length targeting sequences. Target site here is the GFP2 sequence chosen for its robust targeting activity. (C) (Upper) Diagram of targeting sequences across the GFP mRNA and ribosome-binding site (RBS). (Lower) Relative expression of GFP of SauCas9 RNP normalized to sgRNA alone for targeting sequences across the GFP reporter. Dashed red line indicates that the sgRNA alone is as efficient as the RNP for GFP repression. (A–C) Bars represent mean ± S.D. (n = 3). (D) Model for observed SauCas9 ssRNA targeting activity. We propose that accessible RNA is cleaved or repressed efficiently while structured and protein-bound RNA is not targeted by SauCas9.