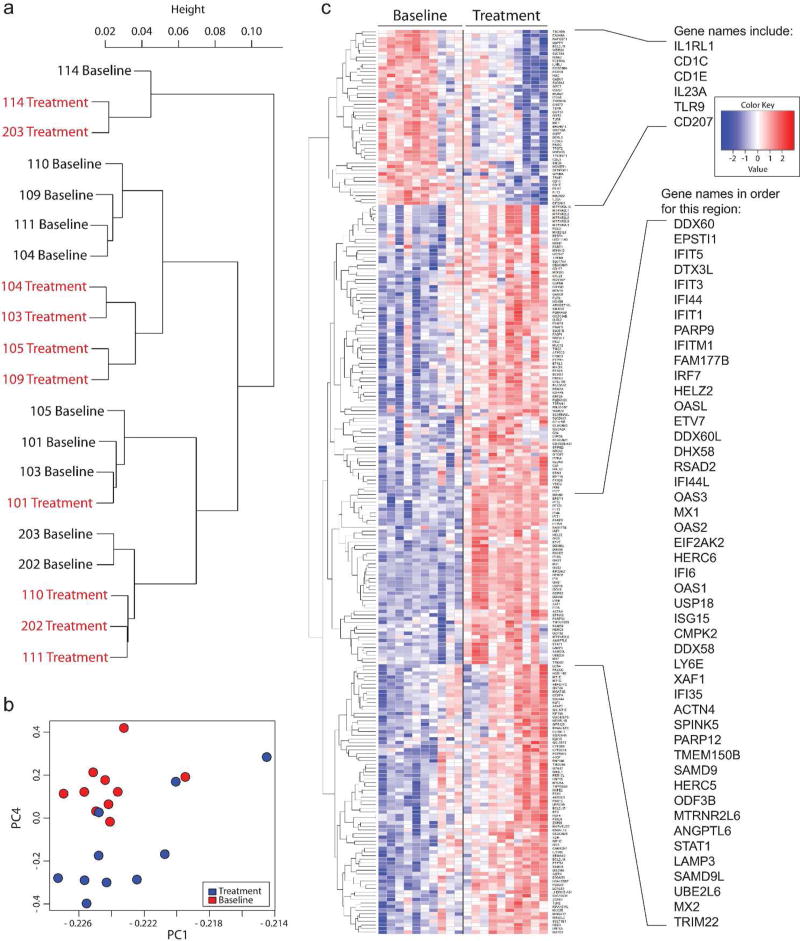
Figure 2. MGN1703 significantly modulated the expression of 248 genes in intestinal mononuclear cells, including many genes involved in type I interferon responses.
(a) Ward link correlation dendrogram for all genes assayed reveals that gene expression patterns cluster with respect to treatment status, rather than by individual (Slatkin-Maddison p=0.0167). (b) Principal component analysis scores plot for PC1 vs. PC4; the two principal components which contributed the maximum variance in the analysis. Plots depicting PC2 and PC3 are presented in Supplemental Figure 1. Samples collected at baseline are represented by red circles and samples collected during treatment are represented by blue circles. (c) RNASeq heat map was generated using standardized Z-scores of the genes that were significantly regulated (up or down) in intestinal mononuclear cells after adjusting for a FDR <0.05. Individual participants are depicted in the columns and one gene is depicted per row. The dendrogram reveals the relatedness of individual gene expression patterns. A full list of significantly regulated genes can be found in Table S3. (n=10 in all graphs)