Abstract
The A118G single nucleotide polymorphism (SNP) of the mu-opioid receptor gene (Oprm1) has been implicated in mediating the rewarding effects of alcohol. Clinical and preclinical studies suggest that the G allele may confer a genetic vulnerability to alcohol dependence, though it remains unknown whether these effects are sex-specific. We used male and female mice homozygous for the “humanized” 118AA or 118GG alleles to determine whether the A118G SNP potentiates ethanol consumption in a sex-specific manner in both the two-bottle choice and drinking-in-the-dark (DID) paradigms. Mice were also assessed for differences in naltrexone sensitivity, ethanol reward assessed via conditioned place preference (CPP), and sensitivity to the sedative/ataxic effects of ethanol using the rota-rod and loss of righting reflex (LORR) assays. We found that male and female 118GG mice drank significantly more ethanol than 118AA littermates using a continuous access, two-bottle choice paradigm. In the limited-access DID drinking model, (i) female (but not male) 118GG mice consumed more ethanol than 118AA mice and (ii) naltrexone pretreatment was equally efficacious at attenuating ethanol intake in both 118AA and 118GG female mice while having no effect in males. Male and female 118GG and female 118AA mice developed a robust conditioned place preference (CPP) for ethanol. Female 118GG mice displayed less sensitivity to the sedative/ataxic effects of ethanol compared to female 118AA mice on both the rota-rod and the LORR assays while male mice did not differ in their responses on either assay. Our findings suggest that increased ethanol consumption in male 118GG mice may be due to increased ethanol reward, while increased drinking in female 118GG mice might be due to decreased sensitivity to the sedative/ataxic effects of ethanol. Collectively, these data might be used to help identify sex-specific pharmacotherapies to combat alcohol use disorders.
Keywords: ethanol, A118G, sex differences, humanized mice, mu-opioid receptor
1. Introduction
Alcohol use disorder is highly prevalent, affecting approximately eight million Americans and inflicting a tremendous cost (in excess of $223.5 billion annually) to society (Grant et al., 2004; Bouchery et al., 2011). Currently, there are only a few federally approved pharmacotherapies available for the treatment of alcohol dependence. This is partially attributable to the differences in clinical efficacies of drugs due to their abilities to work only in specific alcoholic subpopulations (Kenna, 2005). One subpopulation of alcoholics that has recently received considerable attention is patients that possess the A118G single nucleotide polymorphism (SNP) within the mu-opioid receptor (Oprm1) gene.
The A118G mutation is the most common Oprm1 SNP, with an alleleic frequency of 40–60% among those of Asian, 15–30% among those of Caucasian, and 1–3% among individuals of African and Hispanic decent (Bergen et al., 1997; Bond et al., 1998; Gelernter et al., 1999; Tan et al., 2003). This SNP results in an asparagine to aspartic acid amino acid substitution (N40D), disrupting a putative glycosylation site in the receptor originally thought to facilitate increased Oprm1 signaling (Bergen et al., 1997; Bond et al., 1998). However, more recent work suggests that the A118G SNP causes a loss of Oprm1 signaling (Zhang et al., 2005; Mague et al., 2009; Huang et al., 2012; Wang et al., 2012; Weerts et al., 2013).
Although some clinical studies have not found an association between the A118G SNP and alcohol dependence (Bergen et al., 1997; Arias et al., 2006; Gelernter et al., 2007; Tidey et al., 2008; Ooteman et al., 2009; Rouvinen-Lagerström et al., 2013), many others suggest that the G allele confers genetic vulnerability to alcohol dependence (Oslin et al., 2003; Bart et al., 2005; Ray and Hutchison, 2004; 2007; Anton et al., 2008; Chamorro et al., 2012; Hendershot et al., 2016). Individuals expressing at least one copy of the G allele have been reported to have a greater risk of developing alcoholism (Bart et al., 2005). Carriers of the G allele report greater feelings of intoxication, stimulation, sedation, and of happiness or euphoria associated with alcohol consumption (Ray and Hutchison, 2004; 2007), exhibit increased alcohol-stimulated dopamine release in the ventral striatum (Ramchandani et al., 2011), and achieve higher breath alcohol concentrations following increased intravenous alcohol administration in a free access paradigm (Hendershot et al., 2016). Finally, alcoholics expressing at least one copy of the G allele show a greater therapeutic response to naltrexone treatment (Oslin et al., 2003; Ray and Hutchison, 2007; Kranzler et al., 2009), including lower relapse rates (Chamorro et al., 2012) and a delay in a return to heavy drinking (Oslin et al., 2003).
The functionally homologous C77G SNP within the mu-opioid receptor of rhesus monkeys also causes greater ethanol consumption and increased sensitivity to the suppressive effect of naltrexone on ethanol drinking in non-human primates (Barr et al., 2007; 2010; Vallender et al., 2010). One strategy to help decipher the role of the A118G SNP in ethanol addiction involves comparing “humanized” mice that are homozygous for the major A and minor G alleles. Humanized male 118GG mice display a four-fold increase in ethanol-stimulated dopamine release in the nucleus accumbens (Ramchandani et al., 2011), drink more ethanol, show a greater sensitivity to naltrexone, and have a reduced reward threshold for ethanol (Bilbao et al., 2015). These results demonstrate that the A118G SNP causes increased ethanol consumption and reward in male mice. However, the effect of the A118G SNP on ethanol-motivated behavior has not been studied in female mice.
The objective of this study was to test the hypothesis that the A118G SNP potentiates ethanol intake and reward in female mice. We also tested the hypothesis that female mice expressing the A118G SNP would be more sensitive to naltrexone. Finally, we assessed sensitivity to the sedative and ataxic effects of ethanol in order to determine whether increased ethanol intake in female 118GG mice might be due to a decreased sensitivity to the effects of ethanol.
2. Materials and Methods
2.1 Subjects
In total, 189 experimentally naïve adult male (N=78) and female (N=111) A118G mice were used. “Humanized” mice were created by replacing exon 1 of the Oprm1 gene with the corresponding human sequence for the major human 118A allele. Site-directed mutagenesis was used to induce the mutant G allele at position 118. Therefore, these mice are genetically identical with the exception of the induced A→G mutation (for more details, see Ramchandani et al., 2011). Mice were back-crossed onto a C57BL/6 background. Mice were individually (drinking studies) or group housed (all other studies) on a 12:12 hour light/dark cycle (lights out at 19:00) with ad libitum access to food and water except where noted. Animal care procedures were conducted in accordance with NIH guidelines for the Care and Use of Laboratory Animals (2011) and with approval from Pennsylvania State University’s Institutional Animal Care and Use Committee (IACUC).
2.2 Genotyping
Mice were genotyped as previously described (Freet et al., 2015). Specifically, ear snips were obtained and sent to Transnetyx (Cordova, TN) for DNA sequencing.
2.3 Drugs
A 100% ethanol stock solution was diluted to 20% (v/v) in 0.9% sterile saline. A dose of 2 g/kg was administered intraperitoneally (IP) in an injection volume of 12.5 ml/kg for the CPP and rota-rod experiments and at 4 g/kg for the loss of righting reflex and blood ethanol concentration (BEC) experiments. Ethanol was diluted in tap water to 20% (v/v) for the Drinking-in-the-Dark (DID) experiment and to 6, 9, 12, 15, and 18% (v/v) in tap water for the two-bottle choice drinking assays. Sucrose was dissolved in tap water for final concentrations of 1.7 and 4.25% (w/v) and quinine hydrochloride was dissolved in tap water to reach final concentrations of 0.03 and 0.10M. Naltrexone hydrochloride (Sigma Aldrich, St Louis, MO) was dissolved in sterile 0.9% saline and administered 30 minutes prior to testing at a dose of 1 mg/kg IP using an injection volume of 10 ml/kg.
2.4 Two-Bottle Choice Drinking
Male and female 118AA and 118GG mice were given continuous two-bottle choice access to both water and ethanol (v/v) in order to assess ethanol intake and preference. Ethanol and water were made available in conical tubes fitted with sipper tubes containing a ball bearing to prevent leakage, and bottle positions were alternated daily to control for side preference. Ethanol was tested in increasing concentrations (6, 9, 12, 15, and 18% v/v) every day for six consecutive days. Bottles were weighed every day and mice were weighed every other day. Ethanol intake was calculated as g/kg/day and preference as percent intake (mLs)/total fluid intake (mLs). Using this same paradigm, mice were also assessed for sucrose (1.70% and 4.25% w/v) and quinine (0.03M and 0.10M) preference with a two-week wash out period between solutions.
2.5 Ethanol Drinking-in-the-Dark (DID) and Naltrexone Testing
Mice were assessed for binge-drinking behavior using a modified DID procedure (Rhodes et al., 2005; Moore et al., 2007). Briefly, 3 hours into the dark cycle, water bottles were removed and replaced with a pre-weighed bottle containing 20% ethanol (v/v). After two hours of ethanol access, ethanol bottles were removed and water bottles were replaced. Ethanol bottles were then weighed and intake was calculated as g/kg/2-hour session. Ethanol intake was recorded across 4 days, and the averages were analyzed to determine differences in ethanol intake.
2.6 Naltrexone Testing
Using the DID procedure (described above), mice were tested for the effect of naltrexone on binge-drinking. Testing days 1 and 2 of drinking were identical to those described above, with the exception that 30 minutes prior to gaining ethanol access, mice were subjected to handling in order to acclimate them to the injection procedure. Using a within subjects, counterbalanced design, mice were injected IP on days 3 and 4 with either 1 mg/kg of naltrexone, or an equal volume of sterile 0.9% saline, 30 minutes prior to gaining ethanol access. The ethanol bottle was weighed both before and after the mice were given two hour access, with all intakes reported in g/kg.
2.7 Conditioned Place Preference (CPP)
Mice were tested in standard 3-chambered place conditioning boxes (Med Associates, St. Albans, VT) that were individually housed in sound-attenuated chambers. The experiment consisted of four phases: habituation (one session), baseline preference testing (one session), conditioning (eight sessions) and conditioned place preference testing (one session). Sessions were scheduled over 11 consecutive days. During the habituation session (Day 1), mice were given 30 minutes to explore all sections of the CPP apparatus. The following day (Day 2), mice were given another 30 minutes of access to the CPP apparatus during which they could explore all chambers to determine their baseline preferences (pre-conditioning scores). Following the baseline preference session, mice were randomly assigned to have drug or saline paired with either the black or white conditioning chambers. During the conditioning phase (Days 3–10), mice were injected with either saline or ethanol (2 g/kg, IP) on alternating days (i.e., days 3,5,7,9 or 4,6,8,10). Immediately following injections, mice were confined to the appropriate conditioning chamber for 5 minutes. The order of saline and ethanol exposure was counterbalanced within groups. After eight total conditioning sessions (4 ethanol and 4 saline), mice were given a 30-minute preference test session (Day 11) in the absence of drug. The amount of time spent in each chamber was recorded, and a CPP was determined by assessing the amount of time (in seconds) mice spent in the drug-paired chamber pre- versus post-conditioning.
2.8 Rota-rod Testing
Mice were tested using a single station accelerating rota-rod (Med Associates, St. Albans, VT). Mice underwent 2 (female) or 3 (male) days of training (to attain a similar baseline) during which each mouse was given six trials on the rota-rod to acclimate itself to walking on the apparatus. After training, mice were given 2 g/kg of ethanol IP and then tested for sensitivity to the ataxic effects of ethanol 10 minutes post-injection using the rota-rod. Mice were given 2 test trials that were averaged. If the difference between the two trials was greater than 20 seconds, the mouse was given a third test trial. The average of all rota-rod trials was used in the final analysis.
2.9 Loss of Righting Reflex (LORR)
Mice were injected with 4 g/kg IP of ethanol. Following loss of righting reflex (typically in under two minutes), mice were placed upside down in an inverted v-shaped trough so that all four paws were up in the air. Mice were measured for the amount of time (in minutes) it took for them to right themselves. Regaining the righting reflex was defined as the mouse being able to successfully right itself 3 times within 30 seconds.
2.10 Blood Ethanol Concentrations (BECs)
Mice were injected IP with a bolus of 4 g/kg of ethanol. Retro-orbital bloods were measured at 60 and 120 minutes post-injection to assess BECs. Blood samples (100 ul) were collected in capillary tubes. Shortly after collection, the tubes were centrifuged, and plasma samples (5 ul) were assessed using an Analox AM 1 analyzer (Analox Instruments LTD, Lunenberg, MA) to determine BECs (mg%). Ethanol concentration was determined with an amperometric oxygen electrode that measures oxygen consumption during the enzymatic oxidation of alcohol to acetaldehyde.
2.11 Data Analysis
In all experiments, male and female mice were tested at different time points (days and sometimes weeks apart) to ensure that male and female mice were age-matched accordingly, as ethanol consumption, preference, discrimination, and activity have been shown to vary as a function of age (Goodrick et al., 1975; Amir, 1978; Crews et al., 2000). Therefore, in all experiments, male and female mice were analyzed in separate analyses. During the two-bottle choice assay, a mixed factorial analysis of variance (ANOVA) was used to assess intake across each of the 6 days of each ethanol, sucrose, or quinine concentration as the within-subjects factor, with genotype as the between-subjects factor. The averages of each of the six days of intake were then plotted and are shown across each concentration. Both the CPP and DID/naltrexone challenge experiments were analyzed using two-way mixed ANOVAs, with genotype as the between subjects factor and conditioning (CPP) and drug (DID/naltrexone challenge) as the within-subjects factor. The amount of time to fall off the rota-rod (seconds) or to regain the righting reflex (minutes) was assessed using an unpaired t-test in both male and female mice. BECs were analyzed using a two-way ANOVA with time and genotype as the between-subjects factors. Bonferroni post hoc tests were performed where appropriate, and in all analyses significance was set at p<0.05.
3. Results
3.1 Two-Bottle Choice Drinking
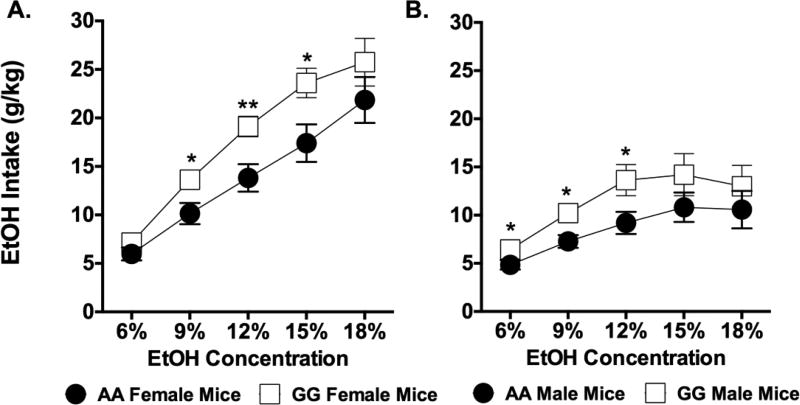
The results of five two-way mixed ANOVAs on two bottle choice drinking revealed significant main effects of genotype on ethanol intake in females at 9% [F(1,38)=6.183, p=0.017], 12% [F(1,38)=8.747, p=0.005], and 15% [F(1,38)=6.222, p<0.001], but not at 6% (p=0.221) or 18% (p=0.419) ethanol. Post hoc analyses showed that female 118GG mice drank considerably more ethanol than female 118AA mice at ethanol concentrations of 9, 12, and 15% (Figure 1A). Ethanol intake was also increased for male 118GG mice at 6% [F(1,13)=4.854, p=0.046], 9% [F(1,13)=7.182, p=0.019], and 12% [F(1,13)=4.959, p=0.044], but not at either 15% (p=0.124) or 18% (p=0.456) ethanol (Figure 1B). There was no effect of genotype on water intake (ml/kg) or ethanol preference (%) in males at any ethanol concentration tested (Table 1). While there was no difference in water intake for females at any ethanol concentration tested, female 118GG mice showed greater ethanol preference compared to 118AA mice at 9 and 12% ethanol (Table 1).
Figure 1. Male and female 118GG mice show increased ethanol consumption in the two-bottle choice assay.
Ethanol intake (g/kg) at each ethanol concentration is shown for both (A) female and (B) male 118GG (GG; open squares) and 118AA (AA; filled circles) mice. Male 118GG (N=5) mice consumed significantly more ethanol (g/kg) than male 118AA (N=10) mice at 6%, 9%, and 12% ethanol. Female 118GG (N=19) mice also drank significantly more ethanol (g/kg) than female 118AA (N=21) control mice at 9%, 12%, and 15%. Data were analyzed using mean ethanol intakes averaged across each of the six days of drinking in two-way mixed ANOVAs. Error bars represent the standard error of the mean (SEM). * = p<0.05; ** = p<0.01.
Table 1.
Ethanol preference (%) and water intake (ml/kg) at each ethanol concentration was assessed using the unlimited access two-bottle choice assay.
| EtOH Preference | 6% | 9% | 12% | 15% | 18% |
|
| |||||
| (%) | |||||
| Female AA | 61.16±2.46 | 62.11±0.76 | 65.24±1.45 | 63.37±1.11 | 61.95±1.29 |
| Female GG | 67.37±3.26 | 76.84±1.71* | 80.56±1.67* | 76.44±1.53 | 64.83±1.17 |
| Male AA | 66.66±3.49 | 70.13±2.50 | 65.31±1.69 | 62.26±2.57 | 53.60±1.71 |
| Male GG | 82.34±1.53 | 82.15±2.02 | 80.15±2.52 | 72.59±2.12 | 58.73±2.38 |
| Water Intake | 6% | 9% | 12% | 15% | 18% |
|
| |||||
| (ml/kg) | |||||
| Female AA | 60.07±3.90 | 65.23±2.55 | 56.09±1.90 | 61.12±2.35 | 70.71±5.03 |
| Female GG | 55.23±5.76 | 43.53±2.82 | 37.23±3.81 | 46.91±4.40 | 76.25±6.42 |
| Male AA | 39.53±4.87 | 34.11±2.59 | 38.72±1.53 | 41.37±2.49 | 52.84±2.89 |
| Male GG | 23.86±2.53 | 21.83±1.58 | 26.00±2.34 | 33.46±3.19 | 53.68±3.49 |
Two-way mixed ANOVAs were performed to analyze genotype differences in ethanol preference and water intake. There were no differences among male 118AA (AA; N=10) or 118GG (GG; N=5) mice in either water intake or ethanol preference. However, female 118GG (N=19) mice did show higher ethanol preference at 9% and 12% ethanol compared to their 118AA (N=21) counterparts. Error bars represent the standard error of the mean (SEM).
= p<0.05 compared to female AA mice.
Results from a two-way ANOVA failed to show a significant main effect of genotype on either sucrose intake (p=0.602) or preference (p=0.537) in female mice (Table 2). There was a significant main effect of sucrose concentration on both intake [F(1,21)=61.62, p<0.001] and preference [F(1,21)=4.895, p=0.0382], with female mice showing an overall greater intake and preference for 4.25% versus 1.7% sucrose. Likewise, no main effects of genotype on either sucrose intake (p=0.258) or preference (p=0.996) were found in male mice. There was a significant main effect of concentration on sucrose intake [F(1,13)=47.05, p<0.001], but not preference (p=0.057). Post hoc analysis revealed that males had a greater intake of sucrose at 4.25% versus 1.70% (Table 2).
Table 2.
(A) Sucrose (g/kg) and (B) quinine (g/kg) intakes and preference (%) at both sucrose and quinine concentrations were assessed using the unlimited access two-bottle choice assay.
| Intake (g/kg) | Preference (%) | |||
|---|---|---|---|---|
| A. Sucrose | 1.70% | 4.25% | 1.70% | 4.25% |
|
| ||||
| Female AA | 3.43±0.38 | 20.88±4.31 | 87.66±1.82 | 89.85±4.79 |
| Female GG | 3.53±0.32 | 23.78±2.76 | 86.97±1.81 | 94.70±0.49 |
| Male AA | 1.83±0.16 | 8.62±1.24 | 83.88±1.34 | 87.90±1.79 |
| Male GG | 2.18±0.35 | 11.82±0.30 | 84.20±1.76 | 87.60±1.36 |
| B. Quinine | 0.03M | 0.10M | 0.03M | 0.10M |
|
| ||||
| Female AA | 0.55±0.03 | 0.74±0.04 | 12.60±0.73 | 15.69±0.80 |
| Female GG | 0.53±0.01 | 0.74±0.04 | 12.85±0.48 | 15.94±0.62 |
| Male AA | 0.56±0.04 | 0.46±0.04 | 15.73±1.07 | 11.99±0.86 |
| Male GG | 0.60±0.14 | 0.49±0.09 | 17.03±4.07 | 12.28±2.05 |
Two-way mixed ANOVAs were performed to analyze genotype differences in (A) sucrose and (B) quinine intakes and preferences. There were no differences among male 118AA (AA; N=10) or 118GG (GG; N=5) mice in intake or preference for sucrose or quinine. Similarly, there were no differences among female 118AA (N=12) or 118GG (N=11) in either intake or preference for sucrose or quinine. Error bars represent the standard error of the mean (SEM).
Two-way ANOVAs failed to find a genotype effect on either quinine intake (p=0.928) or preference (p=0.752) in female mice. However, there was an effect of quinine concentration on both intake [F(1,21)=46.89, p<0.001] and preference [F(1,21)=31.39, p<0.001] with post hoc analyses indicating that female mice preferred and consumed more quinine at a concentration of 0.1M versus 0.03M. There was no effect of genotype on either quinine intake (p=0.664) or preference (p=0.739) in male mice. And while there was not a significant main effect of concentration on quinine intake (p=0.053), there was a significant main effect of concentration on quinine preference [F(1,13)=13.25, p=0.003] such that male mice had a greater preference for 0.03M over 0.10M quinine (Table 2).
3.2 Ethanol Drinking-in-the-Dark (DID) and Naltrexone testing
A two-way mixed ANOVA determined that there were significant main effects of both genotype [F(1,19)=5.599, p<0.029] and naltrexone treatment [F(1,19)=102.500, p<0.001] on ethanol intake in the DID paradigm. However, there was not a significant drug × genotype interaction (p=0.442). Even so, bonferroni post hoc tests showed that female 118GG mice given saline consumed more ethanol during the two hours of DID access than 118AA mice given saline. Treatment with 1 mg/kg naltrexone reduced ethanol intake in both female 118AA and 118GG mice (see Figure 2). Among male mice, there was neither a significant main effect of genotype (p=0.908) or naltrexone treatment (p=0.232), nor a reliable interaction (p=0.272).
Figure 2. 118AA and 118GG female mice are equally sensitivity to 1 mg/kg of naltrexone in a two-hour binge-drinking paradigm.
Ethanol intake (g/kg) was measured using the two-hour limited-access drinking-in-the-dark (DID) paradigm in female 118AA (AA; N=11) and 118GG (GG; N=10) and in male 118AA (N=8) and 118GG (N=12) mice. White bars represent AA mice and grey bars indicate GG mice. Following 4 days of baseline DID drinking, mice were challenged with 1 mg/kg naltrexone (patterned bars) or saline (open bars). Female GG mice drank significantly more ethanol than AA mice following pretreatment with saline while there was no genotype difference in DID drinking for male mice. Naltrexone significantly attenuated ethanol intake (g/kg) in\\ both female AA and GG mice but not in any of the male mice. Data were analyzed using two-way mixed ANOVAs; error bars represent the standard error of the mean (SEM). # = p<0.05; *** = p<0.001.
3.3 Conditioned Place Preference (CPP)
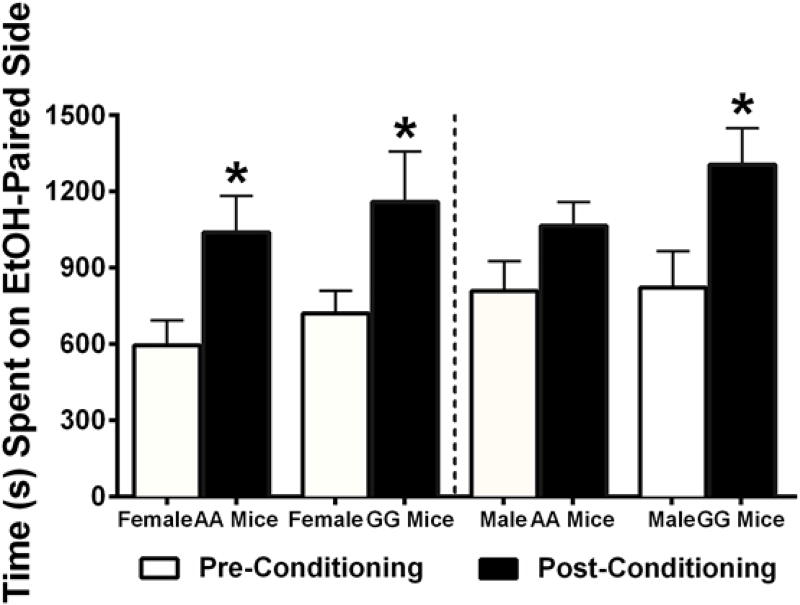
A two-way ANOVA revealed there was an effect of conditioning [F(1,14)=17.53, p<0.001], but not of genotype (p=0.469) or an interaction, (p=0.979) on CPP for ethanol among female mice. Post hoc analyses revealed that both 118AA and 118GG females developed a CPP to 2 g/kg of ethanol following eight days of conditioning. Male mice also showed a significant main effect of conditioning [F(1,14)=24.75, p<0.001] but not of genotype (p=0.450) or an interaction of the two (p=0.152). Post hoc tests revealed that 118GG male but not 118AA male mice developed an ethanol CPP (Figure 3).
Figure 3. Female 118AA and 118GG and Male 118GG mice develop a conditioned place preference (CPP) to the rewarding effects of 2 g/kg of ethanol.
The rewarding effects of 2 g/kg of ethanol were assessed in male and female mice using CPP. The amount of time (seconds) mice spent on the ethanol-paired side both prior to (pre; open bars) and after (post; black bars) ethanol conditioning is shown. Female 118AA (AA; N=8) and 118GG (GG; N=8) and male 118GG (N=8) mice spent more time (in seconds) in the ethanol vs. saline-paired conditioning chamber. In contrast, male 118AA (N=8) mice failed to develop an ethanol CPP. Error bars represent the standard error of the mean (SEM). * = p < 0.001 between pre and post conditioning sessions.
3.4 Rota-rod
Baseline rota-rod performance did not differ between female 118AA and 118GG or between male 118AA and 118GG mice following either 2 (female) or 3 (male) days of training (data not shown). Results from a t-test (Figure 4) showed that 118GG female mice spent significantly more time on the rota-rod after a challenge dose of 2 g/kg of ethanol compared to female 118AA mice [t(16)=2.703, p=0.016]. In contrast, male 118AA and 118GG mice did not differ in the amount of time spent on the rota-rod (p=0.944) following treatment with 2 g/kg of ethanol.
Figure 4. Female 118GG mice are less sensitive to the ataxic effects of 2 g/kg of ethanol on the rota-rod than female 118AA mice.
The amount of time (in seconds) mice spent on the rota-rod following treatment with 2 g/kg ethanol was assessed in female 118AA (AA; N=9) and 118GG (GG; N=9) and male 118AA (N=6) and 118GG (N=7) mice. Female 118GG mice spent more time on the rota-rod compared to female 118AA mice while there was no difference in time spent on the rota-rod for male 118AA and 118GG mice. Error bars represent the standard error of the mean (SEM). * = p<0.05.
3.5 Loss of Righting Reflex (LORR)
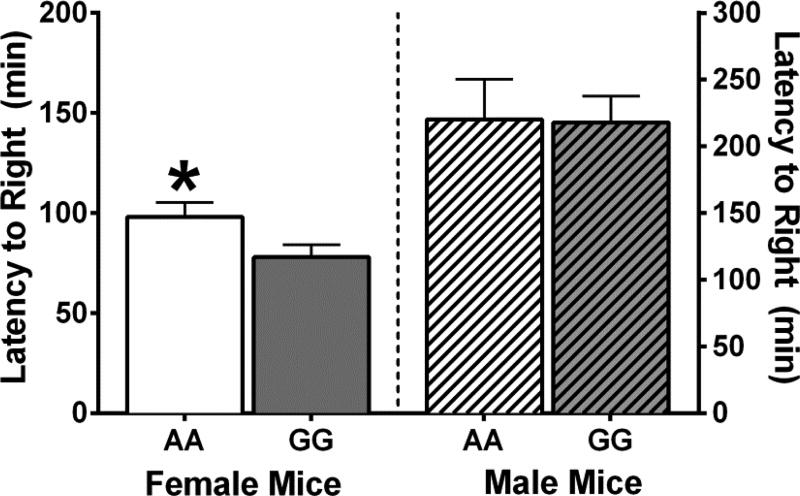
Recovery of the righting reflex was also measured to assess ethanol-induced sedation. Student’s t-tests revealed that while there was no difference in the amount of time it took 118AA and 118GG males to recover their righting reflexes following a challenge dose of 4 g/kg of ethanol [t(12)=0.068; p=0.947], female 118GG mice recovered their righting reflex faster than female 118AA mice [t(16)=2.139; p=0.048; Figure 5]. Interestingly, female mice recovered their righting reflexes (AA=98.12 ±7.11; GG=77.99±6.16 minutes) much faster than male mice (AA=220.30±30.04; GG=217.90±19.81 minutes) by approximately 130 minutes [t(30)=8.601; p<0.001].
Figure 5. Female 118GG mice are less sensitive to the sedative effects of 4 g/kg of ethanol than female 118AA mice.
The amount of time (minutes) to regain the loss of righting reflex (LORR) was assessed following an injection of 4 g/kg of ethanol. Female 118AA (AA; N=9) mice took longer to regain their righting reflex (RR) than female 118GG (GG; N=9) mice. However, there was no difference in the amount of time it took male 118AA (N=5) and male 118GG (N=9) mice to regain their RR. Error bars represent the standard error of the mean (SEM). * = p<0.05.
3.6 Blood Ethanol Concentrations (BECs)
Two-way ANOVAs failed to find any genotype differences in blood ethanol concentrations at either 60 or 120 minutes following a bolus injection of 4 g/kg ethanol in either female [F(1,21)=1.253, p=0.276] or male [F(1,13)=0.557, p=0.469] mice.
4. Discussion
The objective of this project was to better understand whether the A118G SNP in the Oprm1 gene influences ethanol intake, reward, and/or sensitivity in a sex-specific manner. Ethanol intake, in both limited (DID) and unlimited access paradigms, naltrexone-sensitivity, CPP, rotarod, and LORR were examined in male and female 118AA and 118GG mice. The results reveal that sex-specific mechanisms may be responsible for increased ethanol drinking in 118GG mice compared to 118AA controls.
Mice homozygous for the 118G allele consumed more ethanol than their 118AA littermate controls, with 118GG males drinking more ethanol at lower (6–12%) concentrations and female 118GG mice drinking more at slightly higher (9–15%) ethanol concentrations (Figure 1). The lack of any genotype differences in water (Table 1), sucrose, or quinine intake (Table 2) suggests that differences in ethanol intake are likely not due to differences in taste or liquid intake, per se. Interestingly, female (but not male) 118GG mice also showed elevated intake using the two-hour limited-access DID procedure. Since male mice showed genotype differences at lower, but not higher, ethanol concentrations, it is not completely unexpected that male 118GG mice do not show increased consumption of 20% ethanol in the limited-access DID paradigm. These findings are consistent with previous work demonstrating that ethanol intake is increased in humanized male 118GG mice in an open (but not limited) access drinking paradigm (Bilbao et al., 2015). However, this is the first work demonstrating that ethanol consumption is also increased in humanized female 118GG mice.
Results from clinical studies are mixed regarding the role of the A118G SNP on alcoholism. Although a recent review finds that the A118G SNP influences various phenotypes of alcoholism (Ray et al., 2012), meta-analyses conflict over whether there is (Zhang et al., 2006) or is not (Arias et al., 2006) an association between the two. Likewise, while some clinical studies show that alcoholics with at least one copy of the G allele are more responsive to naltrexone treatment (Oslin et al., 2003; Bart et al., 2005; Arias et al., 2006; Chamorro et al., 2012) others find that they are not (Gelernter et al., 2007; Tidey et al., 2008; Oslin et al., 2015; Ziauddeen et al., 2016). Clinically, naltrexone has been reported to reduce the pleasurable effects of alcohol (Swift et al., 1994; Volpicelli et al., 1992; 1995; Ray and Hutchison, 2007), which may be the mechanism by which naltrexone has been shown to lower relapse rates (Chamorro et al., 2012) and delay a return to heavy drinking (Oslin et al., 2003).
Consistent with these clinical studies, Bilbao and colleagues (2015) found that naltrexone attenuated ethanol intake in a free access drinking paradigm in male 118GG (but not 118AA) mice. Therefore, we expected to see an attenuation of ethanol intake in 118GG male mice following naltrexone pretreatment. As female mice were not previously examined, we were unsure how the A118G SNP would alter their responsiveness to naltrexone. Surprisingly, we found that pretreatment with 1 mg/kg of naltrexone attenuated two-hour (limited access DID) ethanol intake in female 118AA and 118GG mice to an equivalent extent, while having no suppressive effect on ethanol consumption in male mice.
Failure to see an effect of naltrexone in the males is somewhat surprising as naltrexone was approved by the FDA for the treatment of alcoholism following the finding that naltrexone-treated patients had significantly better drinking outcomes than their placebo-treated counterparts (O’Malley et al., 1992; Volpicelli et al., 1992). Results of subsequent meta-analyses revealed that clinically, treatment with naltrexone results in a small, modest positive effect compared to placebo, most notably in reducing relapse to heavy drinking (Kranzler & Van Kirk, 2001; Srisurapanont & Jarusuraisin, 2005; Pettinati et al., 2006). It is possible that the males, specifically the 118GG mice, did not consume enough ethanol for naltrexone to have a discernable effect as C57BL/6 mice typically consume in excess of 4 g/kg of ethanol using a similar DID paradigm (Rhodes et al., 2005). Likewise, our mice only had two hours of ethanol access on testing days following naltrexone treatment instead of 4 hours of access, which has been shown to further increase drinking levels (7.5 g/kg; Rhodes et al., 2005, for a review see Thiele & Navarro, 2014). Providing 4 hours of drinking access on testing days would have made it easier to identify an effect of naltrexone on ethanol intake, particularly in males. These concerns are somewhat mitigated given that we were able to find an effect of naltrexone in females displaying similar intakes.
Failure to see an effect of naltrexone in the males or a difference in naltrexone sensitivity in the females could also be due to the dose of naltrexone administered and the involvement of other opioid receptors in mediating ethanol reinforcement. Specifically, while lower doses of naloxone and naltrexone (<1.0 mg/kg) have been shown to preferentially bind to the MOR (Paterson et al., 1984), doses at or exceeding 1.0 mg/kg tend to be considered non-selective as binding also occurs at kappa and delta opioid receptors (Childers et al., 1979; Paterson et al., 1984; Takemori & Portoghese 1984; Emmerson et al., 1994; Wang et al., 2001; 2007).
Our finding of increased ethanol intake in 118GG mice supports both clinical (Oslin et al., 2003; Bart et al., 2005; Ray and Hutchison, 2004; 2007; Anton et al., 2008; Chamorro et al., 2012; Hendershot et al., 2016) and basic research studies suggesting that the G allele may confer a genetic predisposition towards alcoholism. Our results also provide additional evidence that the endogenous opioid system (EOS) is involved in mediating ethanol reinforcement (for a review s2e Herz, 1997), likely through modulation of dopamine release along the mesolimbic dopamine pathway (for a review, see Koob 1992). In this pathway, dopaminergic processes originating in the ventral tegmental area (VTA) give rise to mesolimbic fibers (Ungerstedt 1971; Simon et al., 1976) that project to structures closely associated with the limbic region, most notably the nucleus accumbens (NAc) shell (Koob, 1992; Spanagel, 2009, for a review see Oswald & Wand, 2004). Along this pathway, ethanol-stimulated dopamine release occurs both directly through receptor activation in the NAc and indirectly through the modulation of GABA interneurons in the VTA that otherwise tonically inhibit dopamine release (Johnson & North, 1992; Gianoulakis, 1996; Wise, 1996; Herz 1997; Margolis et al., 2003; Fichna et al., 2007). Dopamine release in the EOS is mediated, at least in part, by the MOR as self-administration of heroin directly into the VTA and systemic administration of morphine facilitated dopamine release in the NAc (Di Chiara and Imperato, 1988; Xi and Stein, 1998). Pretreatment with the MOR antagonist, beta-funaltrexamine, blocked morphine-stimulated dopamine release in the NAc (Di Chiara and Imperato, 1988). Similarly, pretreatment with the opiate antagonist naltrexone can block beta-endorphin and enkephalin-induce increases in dopamine release in the NAc (Koob, 1992; Gonzales and Weiss, 1998). Likewise, MOR knockout mice do not self-administer ethanol in either operant or two-bottle choice paradigms, further confirming the importance of the MOR in the reinforcing aspects of ethanol (Roberts et al., 2000).
Our finding that 118GG (but not 118AA) male mice develop a conditioned place preference for 2 g/kg of ethanol suggests that increased ethanol consumption in male 118GG mice may be due to the enhancement of the rewarding effects of ethanol. This finding is consistent with preclinical work showing that humanized 118GG male mice show a decreased threshold for brain stimulation reward compared to 118AA mice at 0.6 g/kg of ethanol using the intracranial self-stimulation (ICSS) model of reinforcement (Bilbao et al., 2015). Interestingly, there were no differences in ICSS thresholds at higher ethanol doses (1–1.7 g/kg) between male 118AA and 118GG mice. This finding fits well with our results showing differences in ethanol consumption at lower, but not higher, ethanol concentrations in males. Likewise, Ramchandani and colleagues (2011) found a four-fold increase in ethanol-stimulated dopamine release within 10 minutes of administration of 2 mg/kg ethanol in the nucleus accumbens of male 118GG mice compared to male 118AA controls (female mice were not tested). Taken together, these findings suggest that differences in ethanol intake in male 118GG mice might be due to differences in sensitivity to ethanol reward. In contrast to males, both 118AA and 118GG female mice developed a CPP for 2 g/kg of ethanol, suggesting that genotype differences in ethanol intake in female mice are unlikely to be due to altered ethanol reward. This observation is consistent with clinical data reporting that males but not females showed a greater activation of dopamine in the ventral striatum following oral alcohol exposure compared to placebo (Urban et al., 2010).
That said, we find that female 118GG mice are less sensitive to the ataxic effects of 2 g/kg on the rota-rod and are much quicker to recover their righting reflex following a bolus injection of 4 g/kg of ethanol than 118AA females. Examination of blood ethanol concentrations (BECs) following the administration of 4 g/kg of ethanol found no difference in BECs between 118GG and 118AA mice of either sex, suggesting that ethanol metabolism is not altered by the A118G SNP. These findings raise the possibility that the increased ethanol consumption in female 118GG mice may be driven by decreased sensitivity to ethanol. In contrast, 118GG male mice do not differ from 118AA males in their sensitivity to the sedative/ataxic effects of ethanol.
Alcoholism is a complicated disorder. In addition to the opioid and dopaminergic systems, other neurotransmitter systems, including the GABAergic, glutamatergic, cannabinoid, serotonergic, noradrenergic, NPY, and CRF/CRH have all been implicated in regulating various aspects of alcohol use disorders (for a review, see Spanagel, 2009). Unfortunately, given the promiscuity of alcohol and the complicated interplay of neurotransmitter systems mediating alcohol’s diverse effects, only a handful of translatable therapeutics have materialized at the clinical level. Consequently, the field has evolved towards more personalized treatment options, partially through the identification of various clinical subpopulations, such as those with the A118G SNP, who may respond preferentially to select pharmacotherapies.
As such, this is the first study examining sex-specific effects of the A118G SNP on ethanol intake, reward, and sensitivity in mice expressing the humanized A118G mutation. We found that homozygous expression of the human G allele increases ethanol consumption in both male and female mice in an unlimited access drinking paradigm. This is consistent with previous basic and clinical research showing that the G allele confers an increased susceptibility to alcohol use disorders. Interestingly, we found that the increased ethanol intake in male and female 118GG mice may be mediated by sex-specific mechanisms such that male 118GG mice display enhanced sensitivity to ethanol reward in the CPP paradigm, while female 118GG mice exhibit decreased sensitivity to the sedative/ataxic effects of ethanol. Our work is the first to suggest that the A118G SNP may modulate ethanol intake in females and is also the first to suggest that the effect of the A118G SNP on alcohol dependence may occur in a sex-specific manner. Elucidation of the differential mechanisms mediating these sex-specific effects could ultimately aid in the facilitation of more clinically efficacious personalized treatment plans for alcohol use disorders and dependency.
Highlights.
118GG Male and female mice drink more EtOH than 118AA mice
Male A118G mice differed in their response to the rewarding effects of EtOH via CPP
Female A118G mice differed in sensitivity to the sedative/ataxic effects of EtOH
Increased EtOH intake in 118GG mice is likely mediated by sex-specific mechanisms
Acknowledgments
Funding Sources: Penn State University of Medicine Department of Anesthesiology & Perioperative Medicine RIF (DMJ; ANH-R); NIH grant DA036385 (DJM) and funds from the Pennsylvania Department of Health using CURE Tobacco Settlement Funds (DJM)
The authors wish to acknowledge Michael DeLory for his technical assistance in analyzing BECs and Michael Zee and Dr. Diane McCloskey for their critical review of this manuscript
Abbreviations
- ANOVA
Analysis of variance
- BECs
blood ethanol concentrations
- CPP
conditioned place preference
- DID
drinking-in-the-dark
- ICSS
intracranial self stimulation
- IP
intraperitoneal
- LORR
loss of righting reflex
- MOR
mu-opioid receptor
- Oprm1
mu-opioid receptor gene
- NAc
nucleus accumbens
- SNP
single nucleotide polymorphism
- VTA
ventral tegmental area
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anton RF, Oroszi G, O’Malley S, Couper D, Swift R, Pettinati H, Goldman D. An evaluation of the mu-opioid receptor (OPRM1) as a predictor of naltrexone response in the treatment of alcohol dependence. Arch Gen Psychiary. 2008;65:135–144. doi: 10.1001/archpsyc.65.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir S. Brain and liver aldehyde dehydrogenase activity and voluntary ethanol consumption by rats: relations to strain, sex, and age. Psychopharmacology. 1978;57:97–102. doi: 10.1007/BF00426964. [DOI] [PubMed] [Google Scholar]
- Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the μ-opioid receptor gene with substance dependence: a meta-analysis. Drug Alcohol Depen. 2006;83:262–268. doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Barr CS, Chen SA, Schwandt ML, Lindell SG, Sun H, Suomi SJ, Heilig M. Suppression of alcohol preference by naltrexone in the rhesus macaque: a critical role of genetic variation at the mu-opioid receptor gene locus. Biol Psychiatry. 2010;67:78–80. doi: 10.1016/j.biopsych.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr CS, Schwandt M, Lindell SG, Chen SA, Goldman D, Suomi SJ, Higley JD, Heilig M. Association of a functional polymorphism in the mu-opioid receptor gene with alcohol response and consumption in male rhesus macaques. Arch Gen Psychiatry. 2007;64:369–376. doi: 10.1001/archpsyc.64.3.369. [DOI] [PubMed] [Google Scholar]
- Bart G, Kreek MJ, Ott J, LaForge KS, Proudnikov D, Pollak L, Heilig M. Increased attributable risk related to a functional mu-opioid receptor gene polymorphism in association with alcohol dependence in central Sweden. Neuropsychopharmacol. 2005;30:417–422. doi: 10.1038/sj.npp.1300598. [DOI] [PubMed] [Google Scholar]
- Bergen AW, Kokoszka J, Peterson R, Long JC, Virkkunen M, Linnoila M, Goldman D. Mu opioid receptor gene variants: lack of association with alcohol dependence. Mol Psychiatry. 1997;2:490–494. doi: 10.1038/sj.mp.4000331. [DOI] [PubMed] [Google Scholar]
- Bilbao A, Robinson JE, Heilig M, Malanga CJ, Spanagel R, Sommer WH, Thorsell A. A pharmacogenetic determinant of mu-opioid receptor antagonist effects on alcohol reward and consumption: evidence from humanized mice. Biol Psychiatry. 2015;77:850–858. doi: 10.1016/j.biopsych.2014.08.021. [DOI] [PubMed] [Google Scholar]
- Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, Gong J, Schluger J, Strong JA, Leal SM, Tischfield JA, Kreek MJ, Yu L. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchery EE, Harwood HJ, Sacks JJ, Simon CJ, Brewer RD. Economic costs of excessive alcohol consumption in the US, 2006. Am J Prev Med. 2011;41:516–524. doi: 10.1016/j.amepre.2011.06.045. [DOI] [PubMed] [Google Scholar]
- Chamorro A-J, Marcos M, Mirón-Canelo J-A, Pastor I, González-Sarmiento R, Laso F-J. Association of μ-opioid receptor (OPRM1) gene polymorphism with response to naltrexone in alcohol dependence: a systemic review and meta-analysis. Addict Biol. 2012;17:505–512. doi: 10.1111/j.1369-1600.2012.00442.x. [DOI] [PubMed] [Google Scholar]
- Crews FT, Braun CJ, Hoplight B, Switzer RC, Knapp DJ. Binge ethanol consumption causes differential brain damage in young adolescent rats compared with adult rats. Alcohol Clin Exp Res. 2000;24:1712–1723. [PubMed] [Google Scholar]
- Childers SR, Cheese I, Snowman AM, Snyder SH. Opiate receptor binding affected differentially by opiates and opioid peptides. Eur J Pharmacol. 1979;55:11–18. doi: 10.1016/0014-2999(79)90142-0. [DOI] [PubMed] [Google Scholar]
- Di Chiara G, Imperato A. Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely mocing rats. Proc Natl Acad Sci USA. 1988;85:5274–5278. doi: 10.1073/pnas.85.14.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerson PJ, Liu MR, Woods JH, Medzihradsky F. Binding affinity and selectivity of opioids at mu, delta, and kappa receptors in monkey brains. J Pharm Exp Ther. 1994;271:1630–1637. [PubMed] [Google Scholar]
- Fichna J, Janecka A, Costentin J, de Rego J-C. The endomorphin system and its evolving neurophysiological role. Pharmacol Rev. 2007;59:88–123. doi: 10.1124/pr.59.1.3. [DOI] [PubMed] [Google Scholar]
- Freet CS, Ballard SM, Alexander DN, Cox TA, Imperio CG, Anosike N, Carter AB, Mahmoud S, Ruiz-Velasco V, Grigson PS. Cocaine-induced suppression of saccharin intake and morphine modulation of Ca(2)(+) channel currents in sensory neurons of OPRM1 A118G mice. Physiol Behav. 2015;139:216–223. doi: 10.1016/j.physbeh.2014.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Gueorguieva R, Kranzler HR, Zhang HP, Cramer J, Rosenheck R, Krystal JH VA Cooperative Study #425 Study Group. Opioid receptor gene (OPRM1, OPRK1, and OPRD1) variants and response to naltrexone treatment for alcohol dependence: results from the VA cooperative study. Alcohol Clin Exp Res. 2007;31:555–563. doi: 10.1111/j.1530-0277.2007.00339.x. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kranzler J, Cubells J. Genetics of two mu opioid gene (OPRM1) exon 1 polymorphisms: population studies and allele frequencies in alcohol and drug-dependent subjects. Mol Psychiatry. 1999;4:476–483. doi: 10.1038/sj.mp.4000556. [DOI] [PubMed] [Google Scholar]
- Gianoulakis C. Implications of endogenous opioids and dopamine in alcoholism: human and basic science studies. Alcohol Alcohol. 1996;31:33–42. [PubMed] [Google Scholar]
- Gonzales RA, Weiss F. Suppression of ethanol-reinforced behavior by naltrexone is associated with attenuation of the ethanol-induced increase in dialysate dopamine levels in the nucleus accumbens. J Neurosci. 1998;18:10663–10671. doi: 10.1523/JNEUROSCI.18-24-10663.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrick CL. Behavioral differences in young and aged mice: strain differences for activity measures, operant learning, sensory discrimination, and alcohol preference. Exp Aging Res. 1975;1:191–207. doi: 10.1080/03610737508257960. [DOI] [PubMed] [Google Scholar]
- Grant BF, Dawson DA, Stinson FS, Chou SP, Dufour MC, Pickering RP. The 12-month prevalence and trends in DSM-IV alcohol abuse and dependence: United States, 1991–1992 and 2001–2002. Drug Alcohol Depend. 2004;74:223–234. doi: 10.1016/j.drugalcdep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Hendershot CS, Claus ED, Ramchandani VA. Association of OPRM1 A118G and alcohol sensitivity with intravenous alcohol administration in young adults. Addict Biol. 2016;21:125–135. doi: 10.1111/adb.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz A. Endogenous opioid system and alcohol addiction. Psychopharmacol. 1997;129:99–111. doi: 10.1007/s002130050169. [DOI] [PubMed] [Google Scholar]
- Huang P, Chen C, Mague SD, Blendy JA, Liu-Chen LY. A common single nucleotide polymorphism A118G of the mu opioid receptor alters its N-glycosylation and protein stability. Biochem J. 2012;441:379–386. doi: 10.1042/BJ20111050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SW, North RA. Opioids excite dopamine neurons by hyperpolarization of local interneurons. J Neurosci. 1992;12:483–488. doi: 10.1523/JNEUROSCI.12-02-00483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna GA. Pharmacotherapy of alcohol dependence: targeting a complex disorder. Drug Discov Today. 2005;2:71–78. [Google Scholar]
- Koob GF. Drugs of abuse: anatomy, pharmacology, and function of reward pathways. Trends Pharmacol Sci. 1992;13:177–184. doi: 10.1016/0165-6147(92)90060-j. [DOI] [PubMed] [Google Scholar]
- Kranzler HR, Tennen H, Armeli S, Chan G, Covault J, Arias A, Oncken C. Targeted naltrexone for problem drinkers. J Clin Psychopharmacol. 2009;29:350–357. doi: 10.1097/JCP.0b013e3181ac5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR, Van Kirk J. Efficacy of naltrexone and acamprosate for alcoholism treatment: a meta-analysis. Alcohol Clin Exp Res. 2001;25:1335–1341. [PubMed] [Google Scholar]
- Mague SD, Isiegas C, Huang P, Liu-Chen LY, Lerman C, Blendy JA. Mouse model of OPRM1 (A118G) polymorphism has sex-specific effects on drug-mediated behavior. Proc Natl Acad Sci USA. 2009;106:10847–10852. doi: 10.1073/pnas.0901800106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis EB, Hjelmstad GO, Bonci A, Fields HL. K-opioid agonists directly inhibit midbrain dopaminergic neurons. J Neurosci. 2003;23:9981–9986. doi: 10.1523/JNEUROSCI.23-31-09981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore EM, Serio KM, Goldfarb KJ, Stepanovska S, Linsenbardt DN, Boehm SL. GABAergic modulation of binge-like ethanol intake in C57BL/6J mice. Pharmaco Biochem Behav. 2007;88:105–113. doi: 10.1016/j.pbb.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council. Guide for the care and use of laboratory animals (8th ed.) Washington, DC: The National Academies Press; 2011. [Google Scholar]
- O’Malley SS, Jaffe AJ, Chang G, Schottenfeld RS, Meyer RE, Rounsaville B. Naltrexone and coping skills therapy for alcohol dependence: a controlled study. Arch Gen Psychiatry. 1992;49:881–887. doi: 10.1001/archpsyc.1992.01820110045007. [DOI] [PubMed] [Google Scholar]
- Ooteman W, Naassila M, Koeter MW, Verheul R, Schippers GM, Houchi H, Daoust M, van der Brink W. Predicting the effect of naltrexone and acamprosate in alcohol-dependent patients using genetic indicators. Addict Biol. 2009;14:328–337. doi: 10.1111/j.1369-1600.2009.00159.x. [DOI] [PubMed] [Google Scholar]
- Oslin DW, Berrettini W, Kranzler HR, Pettinati H, Gelernter J, Volpicelli JR, O’Brien CP. A functional polymorphism of the mu-opioid receptor gene is associated with naltrexone response in alcohol-dependent patients. Neuropsychopharmacol. 2003;28:1546–1552. doi: 10.1038/sj.npp.1300219. [DOI] [PubMed] [Google Scholar]
- Oslin DW, Leong SH, Lynch KG, Berettini W, O’Brien CP, Gordon AJ, Rukstalis M. Naltrexone vs placebo for the treatment of alcohol dependence: a randomized clinical trial. JAMA Psychiatry. 2015;72:430–437. doi: 10.1001/jamapsychiatry.2014.3053. [DOI] [PubMed] [Google Scholar]
- Oswald LM, Land GS. Opioids and alcoholism. Physiol Behav. 2004;81:339–358. doi: 10.1016/j.physbeh.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Paterson SJ, Corbett AD, Gillan MGC, Kosterlitz HW, McKnight AT, Robson LE. Radioligands for probing opioid receptors. J Recept Res. 1984;4:143–154. doi: 10.3109/10799898409042545. [DOI] [PubMed] [Google Scholar]
- Pettinati JM, O’Brien CP, Rabinowitz AR, Wortman SM, Oslin DW, Kampman KM, Dackis CA. The status of naltrexone in the treatment of alcohol dependence. Specific effects on heavy drinking. J Clin Psychopharmacol. 2006;26:610–625. doi: 10.1097/01.jcp.0000245566.52401.20. [DOI] [PubMed] [Google Scholar]
- Ramchandani VA, Umhau J, Pavon FJ, Ruiz-Velasco V, Margas W, Sun H, Damadzic R, Eskay R, Schoor M, Thorsell A, Schwandt ML, Sommer WH, George DT, Parsons LH, Herscovitch P, Hommer D, Heilig M. A genetic determinant of the striatal dopamine response to alcohol in men. Mol Psychiatry. 2011;16:809–817. doi: 10.1038/mp.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray LA, Barr CS, Blendy JA, Oslin D, Goldman D, Anton RF. The role of the Asn40Asp polymorphism of the mu opioid receptor gene (OPRM1) on alcoholism etiology and treatment: a critical review. Alcohol Clin Exp Res. 2012;36:385–394. doi: 10.1111/j.1530-0277.2011.01633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray LA, Bujarski S, MacKillop J, Courtney KE, Monti PM, Miotto K. Subjective response to alcohol among alcohol-dependent individuals: effects of the mu-opioid receptor (OPRM1) gene and alcohol sensitivity. Alcohol Clin Exp Res. 2013;37:E116–E124. doi: 10.1111/j.1530-0277.2012.01916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE. A polymorphism of the mu-opioid receptor gene (OPRM1) and sensitivity to the effects of alcohol in humans. Alcohol Clin Exp Res. 2004;28:1789–1795. doi: 10.1097/01.alc.0000148114.34000.b9. [DOI] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE. Effects of naltrexone on alcohol sensitivity and genetic moderators of medication response. Arch Gen Psychiatry. 2007;64:1069–1077. doi: 10.1001/archpsyc.64.9.1069. [DOI] [PubMed] [Google Scholar]
- Rhodes JS, Best K, Belknap JK, Finn DA, Crabbe JC. Evaluation of a simple model of ethanol drinking to intoxication in C57BL/6J mice. Physiol Behav. 2005;84:53–63. doi: 10.1016/j.physbeh.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Mcdonald JS, Heyser CJ, Kieffer BL, Matthes HWD, Koob GF, Gold LH. Mu-opioid receptor knockout mice do not self-administer alcohol. J Pharmacol Exp Ther. 2000;293:1002–1008. [PubMed] [Google Scholar]
- Rouvinen-Lagerström N, Lahti J, Alho J, Kovanen L, Aalto M, Partonen T, Silander K, Sinclair D, Räikkönen K, Eriksson JG, Palotie A, Koskinen S, Saarikoski ST. Mu-opioid receptor gene (OPRM1) polymorphism A118G: Lack of association in finnish populations with alcohol dependence or alcohol consumption. Alcohol Alcohol. 2013;48:519–525. doi: 10.1093/alcalc/agt050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon EJ, Hiller Jm, Edelman I. Stereospecific binding of the potent narcotic analgesic [3H]etorphine to rat brain homogenate (opiate/receptor/morphine/antagonist) Proc Nat Acad Sci USA. 1976;70:1947–1949. doi: 10.1073/pnas.70.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanagel R. Alcoholism: a systems approach for molecular physiology to addictive behavior. Physiol Rev. 2009;89:649–705. doi: 10.1152/physrev.00013.2008. [DOI] [PubMed] [Google Scholar]
- Srisurapanont M, Jarusuraisin N. Naltrexone for the treatment of alcoholism: a meta-analysis of randomized controlled trials. Int J Neuropsychopharmacol. 2005;8:267–280. doi: 10.1017/S1461145704004997. [DOI] [PubMed] [Google Scholar]
- Swift RM, Whelihan W, Kuznetsov O, Buongiorno G, Hsuing H. Naltrexone-induced alterations in human ethanol intoxication. Am J Psychiatry. 1994;151:1463–1467. doi: 10.1176/ajp.151.10.1463. [DOI] [PubMed] [Google Scholar]
- Takemori AE, Portoghese PS. Comparative antagonism by naltrexone and naloxone of mu, kappa, and delta agonist. Eur J Pharmacol. 1984;104:101–104. doi: 10.1016/0014-2999(84)90374-1. [DOI] [PubMed] [Google Scholar]
- Tan E-C, Tan CH, Karupathivan U, Yap EP. Mu opioid receptor gene polymorphism and heroin dependence in Asian populations. Neuroreport. 2003;14:569–572. doi: 10.1097/00001756-200303240-00008. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Navarro M. “Drinking in the dark” (DID) procedures: A model of binge-like ethanol drinking in non-dependent mice. Alcohol. 2014;48:235–241. doi: 10.1016/j.alcohol.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidey JW, Monti PM, Rohsenow DJ, Gwaltney CJ, Miranda R, McGeary JE, MacKillop J, Swift RM, Abrams DB, Shiffman S, Paty JA. Moderators of naltrexone’s effects on drinking, urge, and alcohol effects in non-treatment seeking heavy drinkers in the natural environment. Alcohol Clin Exp Res. 2008;32:58–66. doi: 10.1111/j.1530-0277.2007.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban NBL, Kegeles LS, Slifstein M, Xu X, Martinez D, Sakr E, Castillo F, Moadel T, O’Malley SS, Krystal J, Abi-Dargham A. Sex differences in striatal dopamine release in young adults after oral alcohol challenge: a positron emission tomography imaging study with [11C]raclopride. Biol Psychiatry. 2010;68:689–696. doi: 10.1016/j.biopsych.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerstedt U. Stereotaxic mapping of the monoamine pathway in the rat brain. Acta Physio Scand. 1971;367:1–48. doi: 10.1111/j.1365-201x.1971.tb10998.x. [DOI] [PubMed] [Google Scholar]
- Vallender EJ, Rüedi-Bettschen D, Miller GM, Platt DM. A pharmacogenetic model of naltrexone-induced attenuation of alcohol consumption in monkeys. Drug Alcohol Depen. 2010;109:252–256. doi: 10.1016/j.drugalcdep.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli JR, Alterman AI, Hayashida M, O’Brien CP. Naltrexone in the treatment of alcohol dependence. Arch Gen Psychiatry. 1992;49:876–880. doi: 10.1001/archpsyc.1992.01820110040006. [DOI] [PubMed] [Google Scholar]
- Volpicelli JR, Watson NT, King AC, Sherman CE, O’Brien CP. Effect of naltrexone on alcohol “high” in alcoholics. Am J Psychiatry. 1995;152:613–615. doi: 10.1176/ajp.152.4.613. [DOI] [PubMed] [Google Scholar]
- Wang D, Raehal KM, Bilsky EJ, Sadee W. Inverse agonist and neutral antagnoists at mu opioid receptor (MOR): possible role of basal receptor signaling in narcotic dependence. J Neurochem. 2001;77:1590–1600. doi: 10.1046/j.1471-4159.2001.00362.x. [DOI] [PubMed] [Google Scholar]
- Wang D, Sun X, Sadee W. Different effects of opioid antagonists on mu-, delta-, and kappa-opioid receptors with and without agonist pretreatment. J Pharm Exp Ther. 2007;321:544–552. doi: 10.1124/jpet.106.118810. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Huang P, Ung A, Blendy JA, Liu-Chen LY. Reduced expression of the mu opioid receptor in some, but not all, brain regions in mice with OPRM1 A112G. Neuroscience. 2012;205:178–184. doi: 10.1016/j.neuroscience.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerts EM, McCaul ME, Kuwabara H, Yang X, Xu X, Dannals RF, Frost JJ, Wong DF, Wand GS. Influence of OPRM1 Asn40Asp variant (A118G) on [11C]carfentanil binding potential: preliminary findings in human subjects. Int J Neuropsychopharmacol. 2013;16:47–53. doi: 10.1017/S146114571200017X. [DOI] [PubMed] [Google Scholar]
- Wise RA. Addictive drugs and brain stimulation reward. Annu Rev Neurosci. 1996;19:319–340. doi: 10.1146/annurev.ne.19.030196.001535. [DOI] [PubMed] [Google Scholar]
- Xi ZX, Stein EA. Nucleus accumbens dopamine release modulation by mesolimbic GABAA receptors –an in vivo electrochemical study. Brain Res. 1998;798:156–165. doi: 10.1016/s0006-8993(98)00406-5. [DOI] [PubMed] [Google Scholar]
- Ziauddeen H, Nestor LJ, Subramaniam N, Dodds C, Nathan PJ, Miller SR, Sarai BK, Maltby K, Fernando D, Warren L, Hosking LK, Waterworth D, Korzeniowska A, Win B, Richards DB, Johnson LV, Fletcher PC, Bullmore ET. Opioid antagonists and the A118G polymorphism in the mu-opioid receptor gene: effects of GSK1521498 and naltrexone in healthy drinkers stratified by OPRM1 genotype. Neuropsychopharmacol. 2016;41:2647–2657. doi: 10.1038/npp.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Luo X, Kranzler HR, Lappalainen J, Yang B-Z, Krupitsky E, Zvartau E, Gelernter J. Association between two mu-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet. 2006;15:807–819. doi: 10.1093/hmg/ddl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang D, Johnson AD, Papp AC, Sadee W. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280:32618–32624. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]