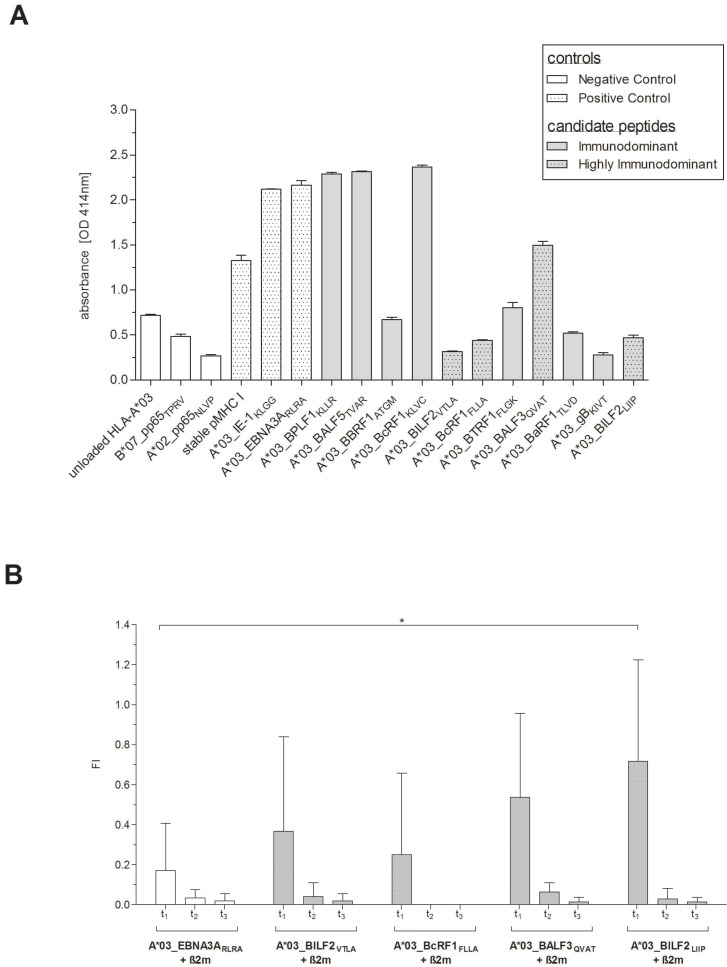
Figure 4.
(A–B) Confirmation of the EBV-derived peptides’ in vitro binding and dissociation. Peptide-binding efficiency on HLA-A*03:01 monomers and stability of the pHLA-A*03:01-complexes of the newly identified EBV-derived candidate-peptides (Table 1) were determined by (A) Flex-TTM MHC Tetramer assays (Flex-T assay) and by (B) T2 peptide binding and dissociation assays. Flex-T assays were carried out for all eleven candidate-peptides. A stable HLA class I monomer with a high-affinity, non UV-labile peptide as well as pMHC monomers for the known HLA-A*03:01-restricted CMV-IE1-specific peptide KLGGALQAK (A*03_IE1KLGG) and A*03_EBNA3ARLRA (Table 1) functioned as positive controls. Unloaded HLA-A*03:01 monomers (unloaded HLA-A*03:01) as well as pMHC monomers for the CMVpp65-derived peptides restricted to HLA-A*02:02 (NLVPMVATV, A*02_pp65NLVP) and HLA-B*07:02 (TPRVTGGGAM, B*07_pp65TPRV), respectively, served as negative controls. (A) Efficiency of the pivotal exchange process was verified by a rapid streptavidin-capture ELISA and results are given as the optical density (OD), quantified at 414 nm absorbance. The stability of the pHLA-A*03:01-complexes in response to the four highly immunodominant EBV-derived peptides was investigated by T2 peptide binding and dissociation assays. Peptide-unloaded T2 cells served as negative control and the known immunodominant HLA-A*03:01-restricted EBV-derived peptide RLRAEAQVK (A*03_EBNA3ARLRA) as reference. (B) The HLA class I expression levels as well as the peptide-MHC-complex dissociation were investigated at the given points of time (t1 = 0 min, t2 = 60 min, t3 = 120 min). The resultant fluorescence index (FI) was calculated as the mean fluorescence intensity (MFI) of HLA-A*03:01 on peptide-stimulated transduced T2 cells and peptide-unloaded cells, respectively. In terms of a comparable reference the unloaded cells were standardized to 0. The results are shown as the mean of n = 3 independent experiments ± SD and statistically significant differences between the EBV-specific peptides and A*03_EBNA3ARLRA are asterisked (*p < 0.05).