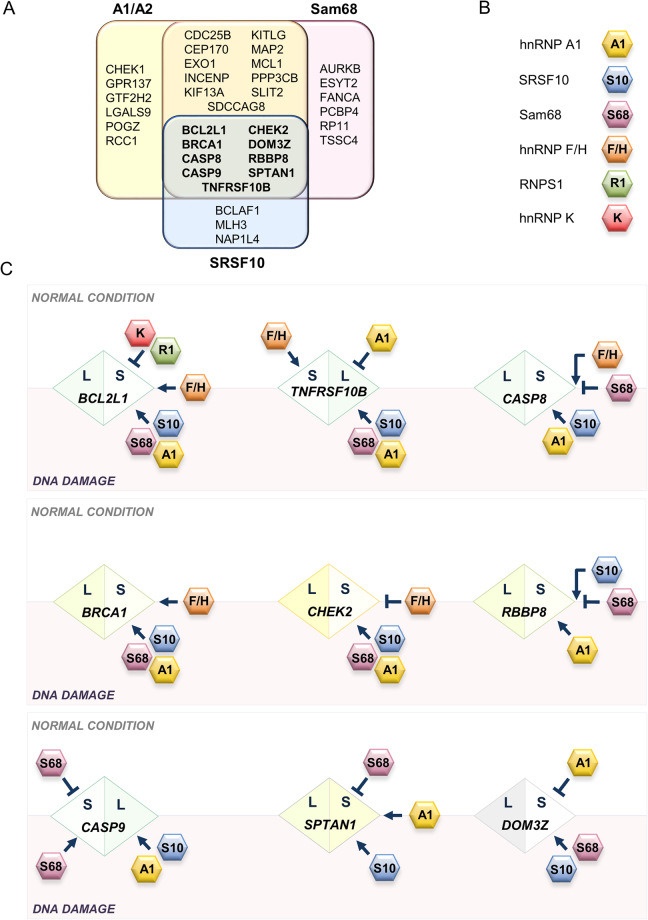
Figure 5.
Control the splicing response to DNA damage by hnRNP A1/A2, Sam68 and SRSF10. (A) Summary of the oxaliplatin-sensitive alternative splicing events (ASE) that were reactive to the depletion of hnRNP A1/A2, Sam68 and SRSF10. ASEs reactive to all three are indicated in bold. (B,C) Diagrams representing splicing outcome and control strategies for the 9 ASEs that are controlled by SRSF10, hnRNP A1/A2 and Sam68 in normal and DNA damage conditions. The diamond contains the name of gene with the S and L half-portions respectively indicating the short and long variants. The direction of the splicing shift upon DNA damage is always from left to right, hence short to long for the CASP9 ASE but long to short for the DOM3Z ASE. RBPs implicated in splicing control are based on this study and reference18. RBPs in the above white area act during normal conditions. RBPs in the gray area are active upon DNA damage. RBPs overlapping both areas contribute to splicing control in both conditions. The impact of RBPs on the production of the L and S variants is indicated. For simplicity, A1 is used to indicate hnRNP A1/A2 proteins.