FIGURE 8.
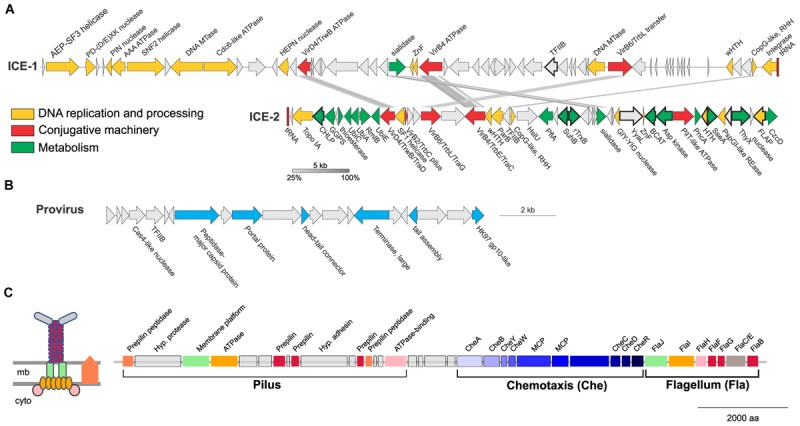
Mobile genetic elements and cell appendages encoded in the genome of Ca. N. cavascurensis. (A) Two putative Integrative Conjugative Elements (ICE) with sequence similarities shown by connections between the two. Genes are colored by function (see legend). CHLP, geranylgeranyl diphosphate reductase; GGPS, geranylgeranyl pyrophosphate synthetase; UbiC, chorismate lyase; UbiA, 4-hydroxybenzoate octaprenyltransferase; RmlB, dTDP-glucose 4,6-dehydratase; UbiE, Ubiquinone/menaquinone biosynthesis C-methylase; HslU, ATP-dependent HslU protease; PflA, Pyruvate-formate lyase-activating enzyme; SuhB, fructose-1,6-bisphosphatase; TrxB, Thioredoxin reductase; YyaL, thoiredoxin and six-hairpin glycosidase-like domains; BCAT, branched-chain amino acid aminotransferase; PncA, Pyrazinamidase/nicotinamidase; SseA, 3-mercaptopyruvate sulfurtransferase; ThyX, Thymidylate synthase; CzcD: Co/Zn/Cd efflux system. (B) A head-tail like provirus from the Caudovirales order with genes in blue indicating putative viral functions. (C) Ca. N. cavascurensis specific type IV pilus locus next to a set of chemotaxis and flagellum (archaellum) genes. Homologs shared between the flagellum and the T4P are displayed with the same colors. A scheme of the putative type IV pilus is shown on the left with the same color code (inspired by drawings in Makarova et al., 2016). mb, membrane; cyto, cytoplasm; MCP, methyl-accepting chemotaxis proteins.
