FIGURE 2.
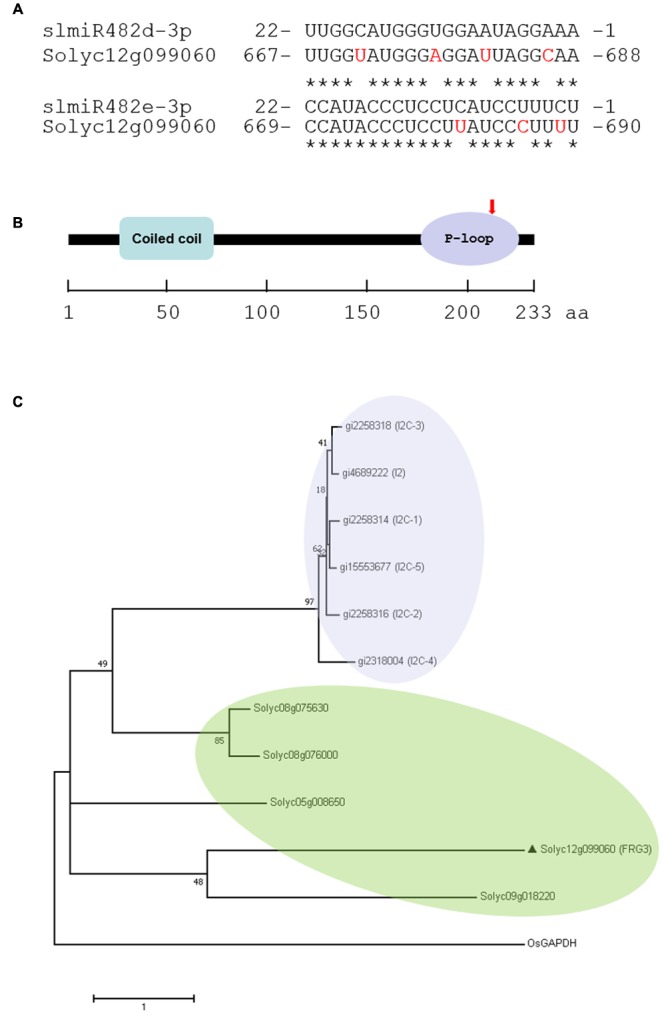
Prediction of slmiRNA targets and protein domain analysis. (A) Predicted mRNA target sequences and targeting site. Targets of slmiR482 family members were identified using the psRNATarget algorithm (http://plantgrn.noble.org/v1_psRNATarget/). Alignments were made using ClustalW (http://www.ebi.ac.uk/Tools/psa/). The nucleotides shown in red in the mRNA are mismatches with the corresponding miRNA. (B) Prediction of translated protein domains. Domain prediction was performed using Interpro (http://www.ebi.ac.uk/interpro/). CC: Coiled-Coil domain. The miRNA binding site is indicated with a red arrow in the P-loop region. (C) Phylogenetic analysis of Solyc12g099060 and I2 homologs. The Maximum Likelihood method based on the JTT matrix-based model. PAM was used for an amino acid transition matrix. OsGAPDH was used as a root by midpoint method. Evolutionary analyses were conducted in MEGA7. The sequences were including gi| 4689222 (I2), gi| 2258314 (I2C-1), gi| 2258316 (I2C-2), gi| 2258318 (I2C-3), gi| 2318004 (I2C-4), gi| 15553677 (I2C-5), Solyc08g075630, Solyc08g076000, Solyc09g018220, Solyc05g008650, Solyc12g099060 (FRG3) and OsGAPDH.
