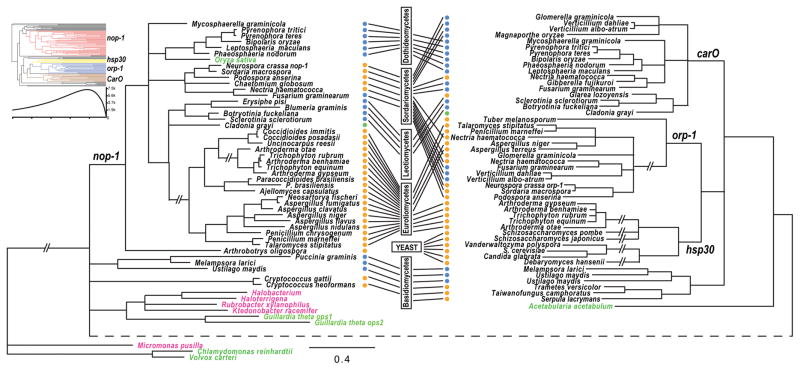
Figure 1.
Phylogeny of fungal opsins and opsin-like proteins inferred from comparing amino acid sequences by maximum likelihood with PhyML under a WAG model. Branches without strong support (bootstrap ≤ 50%) were collapsed. Taxa for opsins from prokaryotic organisms (pink). Taxa for opsins from green algae and other plants (green). Taxon ecology indicated by dots: blue (plant associated), green (lichens) and orange (saprotrophic/animal pathogens). Boxes and lines indicate current fungal classification of these taxa into six major fungal groups. The double slash symbols (—//—) indicate extremely long internodes shortened to efficiently display the tree. The dashed internode across the bottom of the figure was stretched to facilitate side-by-side comparison of the nop-1 clade with the other three clades. Top-left: phyloinformativeness of fungal opsin-like protein was measured and presented with color-coded four clades, nop-1 (red), hsp30 (yellow), orp-1 (blue) and CarO (brown), in an ultrametric phylogeny. High phylogenetic informativeness during recent evolution is consistent with the poor resolution of the phylogeny among early lineages of fungal opsins.