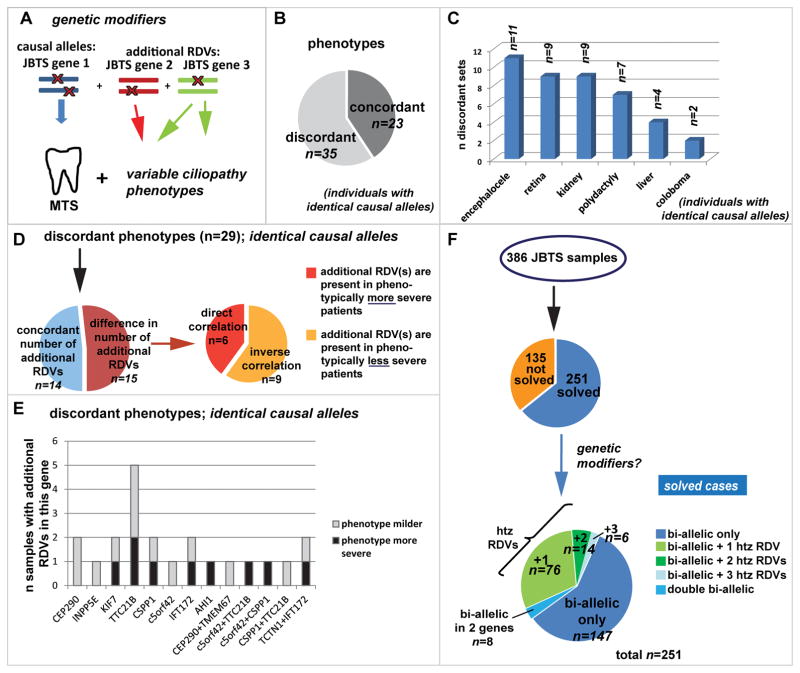
Figure 4. Investigating genetic modifiers among JBTS genes in Joubert syndrome.
(A) Genetic modifiers: Mutations in JBTS gene 2 and 3 modify the phenotypic outcome of the disorder, which is caused by bi-allelic mutations in JBTS gene 1. (B) Pie-chart indicating the proportion of affected individuals sharing identical causal RDVs but for whom phenotypic features are discordant. (C) Bar graph showing the discordant phenotypic features present or not in addition to the Molar Tooth Sign in individuals with shared causal RDVs. (D) The left pie chart indicates for 29 sets of affected individuals with shared causal alleles but discordant phenotypes, the proportion with concordant (blue) or discordant (red) numbers of additional RDVs. Note that phenotypic discordance was observed despite concordant numbers of additional RDVs 50% of the time. The right pie chart shows for those individuals with discordant phenotypes AND discordant RDV numbers (n=15), the proportion of patients in whom the additional RDVs correlated with more severe and with less severe phenotypes. (E) For individuals with shared causal alleles: Bar graph indicating the number of instances where presence of additional RDVs in each of the displayed genes corresponded with a milder (grey) or with a more severe (black) phenotype than in individuals lacking these additional RDVs. (F) Considering the entire JBTS cohort (n=386 individuals), we identified additional heterozygous (htz) RDVs in 96/251(=38%) “genetically solved” individuals with JBTS. Eight affected individuals carried two RDVs in each of two different genes.