Abstract
microRNAs (miRNAs) are small gene regulatory RNAs, and their expression has been found to be dysregulated in a number of human diseases. To facilitate the discovery of small molecules capable of selectively modulating the activity of a specific miRNA, we have utilized new high-throughput screening technology targeting Dicer-mediated pre-miRNA maturation. Pilot screening of ~50,000 small molecules and ~33,000 natural product extract libraries against pre-miR-21 processing indicated the potential of our assay for this goal, yielding a campaign Z′ factor of 0.52 and an average plate signal-to-background (S/B) ratio of 13. Using two-dimensional screening against a second pre-miRNA, pre-let-7d, we evaluated the selectivity of confirmed hits. The results presented demonstrate how high-throughput screening can be used to identify selective small molecules for a target RNA.
Keywords: microRNAs, Dicer, cat-ELCCA, high-throughput screening
Introduction
microRNAs (miRNAs) are a class of >2000 ~22-nucleotide (nt) RNAs important in the posttranscriptional regulation of gene expression.1,2 These small RNAs function by binding to sequence complementary sites in the 3′ untranslated region (UTR) of target genes and inhibiting protein synthesis. Over the past three decades since the discovery of these micromanagers, miRNAs have been shown to regulate the expression of the majority of human genes, including many of those relevant to health and disease.2 Aberrant expression of miRNAs has been linked to the development of most human diseases, including cancer and cardiovascular, viral, and neurodegenerative diseases.2,3 Thus, strategies that will enable modulation of a miRNA’s level in the cell could have enormous therapeutic potential. Although antisense oligonucleotide (ASO)–based approaches have traditionally been utilized for such efforts,3 many challenges still exist in the development of ASO-based reagents for clinical use.2,4 These roadblocks highlight the importance of discovering selective small-molecule ligands for RNA targets, including miRNAs, as an alternative strategy for drugging this class of biomolecules.
The biogenesis of a mature miRNA is derived from two hairpin loop intermediates: a nuclear pri-miRNA (≥1000 nt) and a cytosolic pre-miRNA (~60–80 nt).1 In the final step of maturation, the RNase III enzyme Dicer cleaves the pre-miRNA to form a mature miRNA duplex, of which one strand is loaded into the RNA-induced silencing complex (RISC) to silence gene expression. Thus, as a biological basis for the targeting of miRNAs, we hypothesize that small molecules able to productively bind to a pre-miRNA will inhibit miRNA maturation and thereby activity. Although both pri- and pre-miRNAs contain significant secondary structure for selective small-molecule ligand binding, the targeting of pre-miRNAs was chosen over that of pri-miRNAs due to their favorable size for assay development and screening, and cellular localization for intracellular small-molecule targeting.
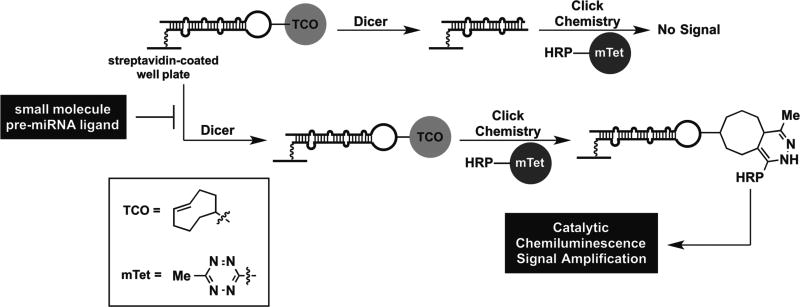
To enable this goal, we have developed new platform assay technology for identifying small-molecule pre-miRNA ligands using our catalytic enzyme-linked click chemistry assay (cat-ELCCA) technology.5–8 In brief, cat-ELCCA is analogous to a chemical enzyme-linked immunosorbent assay (ELISA), whereby click chemistry is employed to enable catalytic signal amplification through covalent conjugation of an immobilized biomolecule (e.g., pre-miRNA) to horseradish peroxidase (HRP) (Fig. 1). For Dicer-mediated pre-miRNA maturation, the assay involves immobilization of a biotinylated pre-miRNA substrate containing a click chemistry handle in the terminal loop. Upon treatment with Dicer, the loop is cleaved, and no chemiluminescence signal is observed; however, if a small-molecule ligand able to inhibit maturation is added, then the loop will be retained and detected. We have previously optimized the click chemistry detection step for application of this pre-miRNA maturation cat-ELCCA to high-throughput screening (HTS).8 The resulting assay exhibited excellent assay statistics, including a signal-to-background (S/B) ratio of 11.5 and Z ′ factor of 0.69,8 indicating its potential in larger screening efforts. In this report, we describe our assay statistics and HTS results after screening ~50,000 small molecules and ~33,000 natural product extracts (NPEs). We also demonstrate how cat-ELCCA can be used to discover selective small-molecule ligands for a pre-miRNA through two-dimensional HTS.
Figure 1.
cat-ELCCA for Dicer-mediated pre-miRNA maturation and identifying small-molecule pre-miRNA ligands.
Materials and Methods
Pre-microRNA and HRP Labeling
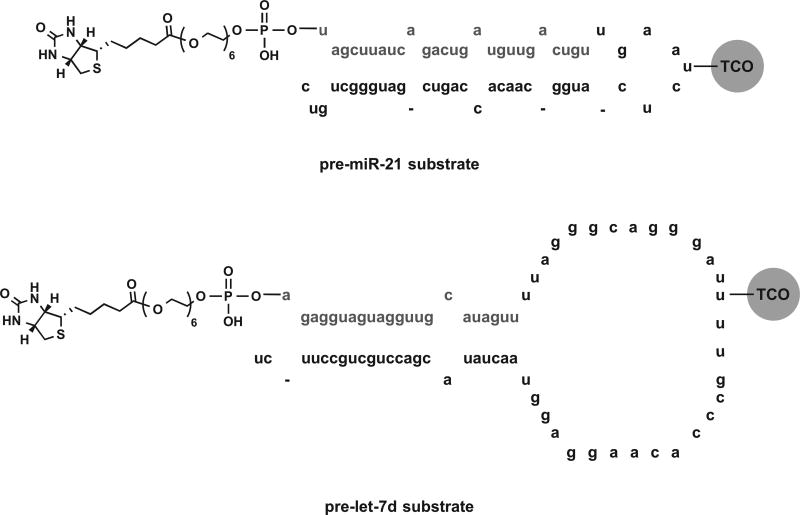
Chemically synthesized pre-miR-21 and pre-let-7d (deprotected, desalted, and high-pressure liquid chromatography [HPLC] purified), containing aminoallyl uridine modifications in the terminal loop and biotin-terminated 18-atom polyethylene glycol (PEG) spacers off of the 5 ′ hydroxyl of the 5 ′ terminal nucleotide (see Fig. 3), were purchased from GE Healthcare Dharmacon (Lafayette, CO) and used as received. Use of labeled pre-miRNA substrates was previously shown to have little to no impact on Dicer processing both in cat-ELCCA and by fluorescent gel analysis.7,8 HRP was purchased from Pierce and used as received. Click handle labeling of the pre-miRNA substrates and HRP was performed as previously described.8 Of note, two separate batches of pre-miR-21 were used for the screen: one for the small molecules and one for the NPEs.
Figure 3.
pre-miRNA substrate structures for primary and two-dimensional screening.
Dicer Expression and Purification
Dicer was prepared as previously reported and stored at −20 °C in 20 mM Tris (pH 7.5), 100 mM NaCl, 1.0 mM MgCl2, 50% glycerol, and 0.1% Triton X-100.8
HTS Assay Protocol
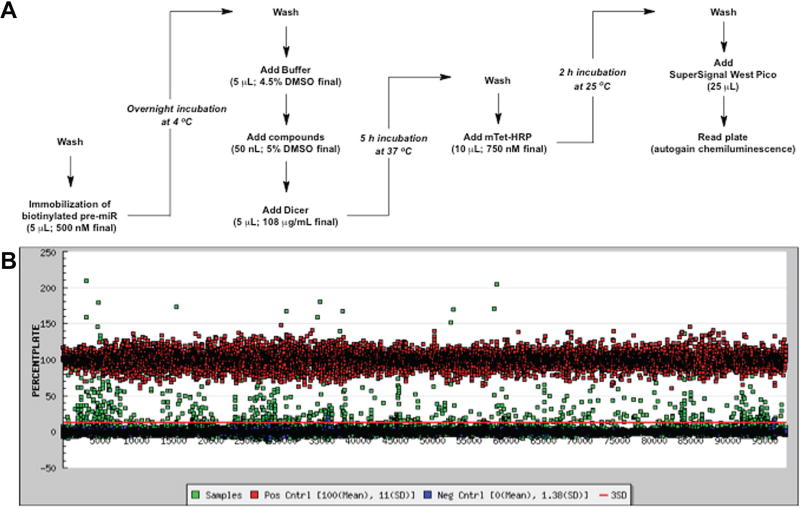
Black, standard-capacity streptavidin-coated 384-well plates (Pierce 15407) were first washed with 50 µL of phosphate buffer (100 mM, pH 7.0; PB7) three times using a Biotek 405 ELX plate washer (Winooski, VT). Subsequently, 5 µL of biotinylated pre-miRNA substrate (500 nM final) was dispensed into the plate using a Multidrop Combi Reagent Dispenser (Thermo Scientific, Waltham, MA). Plates were then centrifuged for 1 min at 1000 rpm (223g), sealed with plate tape, and incubated overnight at 4 °C. The following morning, plates were washed three times with 50 µL of PB7, followed by the addition of 5 µL of Dicer digest buffer (20 mM Tris, 12 mM NaCl, 2.5 mM MgCl2, 1 mM fresh DTT, and 4.5% DMSO)7,8 and centrifugation. Compounds (50 nL of 5 mM DMSO stock, 25 µM final) were then added into the sample wells using a Sciclone (Caliper, Hopkinton, MA) liquid handler with a V&P pin-tool (San Diego, CA); the same volume of DMSO was added to the control wells. The plates were incubated at 25 °C for 15 min before addition of 5 µL of digest buffer containing 217 µg/nL Dicer (108 µg/mL Dicer, 5% glycerol, and 0.01% Triton X-100 final; excess with respect to pre-miRNA). For the positive control wells, digest buffer without Dicer was added. The plates were centrifuged again and resealed before being placed in a 37 °C incubator for 5 h. After Dicer cleavage, plates were washed three times with 50 µL of PB7. mTet-HRP in PB7 (10 µL, 750 nM final) was then dispensed into each well. The plates were subsequently centrifuged, sealed, and incubated at 25 °C for 2 h. Plates were then washed three times with 50 µL of wash buffer (2 mM imidazole, 260 mM NaCl, 0.5 mM EDTA, 0.1% Tween-20 [pH 7.0]), followed by washing three additional times with 50 µL of PB7. Finally, SuperSignal West Pico (25 µL; Pierce) was added, the plates were incubated at 25 °C for 5 min, and chemiluminescence signal was detected using a PHERAstar plate reader using the LUM plus module (BMG Labtech, Cary, NC).
Compound Libraries
Compounds screened were housed at the University of Michigan Center for Chemical Genomics (CCG). For the primary screen, 47,130 compounds from the following collections were used: Sigma LOPAC library of pharmacologically active compounds (1280), Prestwick library of approved drugs (1280), ChemDiv 100K library (21,120), Maybridge MB24K library (23,552), and University of Michigan Chemistry library (895). Additionally, a library of 32,301 NPEs was also tested.9 Compounds were tested at 25 µM in the primary and confirmation screens using 5 mM DMSO stocks. Concentration–response curves (CRCs) were generated over eight points (1.67-fold serial dilution) from 3.3 to 120 µM using 5 mM DMSO stocks; however, compounds were first dispensed with a Mosquito ×1 (TTP Labtech, Cambridge, MA) into polypropylene 384-well plates (Greiner 784201, Monroe, NC), and subsequently diluted with Dicer digest buffer (15 µL) before addition of diluted compound into the pre-miRNA-immobilized plate (5 µL). NPEs were tested at 75 µg/mL in the primary and confirmation screens using 15 mg/mL stocks.
Assay Performance
To monitor assay sensitivity and evaluate its robustness, S/B ratios and Z′ factors were calculated as previously described.10
Data Analyses
HTS data were monitored and analyzed using MScreen.11 Small molecules were considered initial hits if they exhibited ≥5% inhibition by plate based on the negative controls. For the NPEs, this threshold was raised to ≥10% inhibition by plate based on the negative controls. Potential hits meeting these criteria (1480 small molecules and 339 NPEs) were confirmed by rescreening in triplicate. Compounds showing inhibition at ≥3 standard deviations (SD) by plate from the negative controls were considered confirmed hits and analyzed in CRCs in duplicate (170), excluding the NPEs, which underwent more stringent analysis to select those for regrowth. Average percent inhibitions by plate at 120 µM (small molecules) and 75 µg/mL (NPEs) were determined from sample and positive control values normalized to the negative control. All data were analyzed using GraphPad Prism version 6.0c for Mac OS X (GraphPad Software, La Jolla, CA, www.graphpad.com).
Results
HTS to Identify Small-Molecule Ligands for pre-miR-21
Using our previously optimized HTS-optimized cat-ELCCA for pre-miRNA maturation,8 we carried out a pilot screen of compound collections housed at the University of Michigan CCG. As a disease-relevant pre-miRNA substrate, we chose pre-miR-21, which is a validated oncogenic miRNA (oncomiR) overexpressed in the majority of human cancers.12–15 Because of the importance of this miRNA, it has been subject to previous probe discovery campaigns by others in the field.16–19 Our assay protocol is summarized in Figure 2A and was performed using 384-well, standard-capacity, black streptavidin-coated well plates. To facilitate the development of cat-ELCCA and pursue the discovery of pre-miR-21-selective binders, we screened a diversity of chemical matter representing pharmacologically active molecules and known drugs (LOPAC, Prestwick; 2560 compounds), small-molecule scaffolds (ChemDiv, Maybridge, University of Michigan Chemistry collection; 45,567 compounds), and NPEs (32,301 mixtures). Purified compounds were tested at 25 µM, while NPEs were tested at 75 µg/mL, based on available stock concentrations, to ensure that weak actives with potential selectivity would be detected. For each plate, 32 negative control (DMSO, Dicer; coefficient of variation [CV] = 9.5%) and 32 positive control (DMSO, no Dicer; CV = 14%) wells were incorporated into columns 1–2 and 23–24, respectively. The assay performed excellently throughout our screening efforts with a campaign Z′ factor of 0.52, average plate Z′ factor of 0.63, and average plate S/B of 13. Because our goal was to discover compounds selective for pre-miR-21, from our primary screen, we used the generous hit criteria of ≥5% and ≥10% inhibition by plate based on the negative controls for the small molecules and NPEs, respectively. Using this cutoff, 1480 small molecules (3.1% hit rate) and 339 NPEs (1.0% hit rate) were identified (Fig. 2B). Of note, 0.01% Triton X-100 was included in the compound incubation and Dicer cleavage steps to decrease the chance of the discovery of small molecules prone to aggregation in biochemical assays.20,21
Figure 2.
HTS of cat-ELCCA for Dicer-mediated pre-miRNA maturation. (A) Detailed assay protocol in 384-well plates. (B) Screening campaign data.
Confirmation of Identified Hits
For confirmation of activity, hit compounds from our primary screen were tested in triplicate. From the small-molecule hits, compounds showing inhibition at ≥3 SD by plate from the negative controls at 25 µM were considered true, resulting in 170 confirmed hits (11.5% of hits). We attribute this significant loss of chemical matter to the fact that many of the retested compounds exhibited very weak inhibitory activity and reactive and pan-assay interference compounds (PAINS) compounds were not removed.22 Additionally, spurious striping was observed on a few plates due to the dispenser. With respect to compound interference, common fluorescent molecules and fluorescence quenchers within the libraries were not found to interfere with the assay, as noted previously.7 For the NPEs, 47 extracts were confirmed showing ≥10% inhibition by plate upon triplicate analysis (14% of hits). Again, compound interference was not observed, even in the several highly fluorescent samples known to interfere in fluorescence-based assays. As the NPEs are all mixtures, it is not surprising that many mixtures did not repeat due to the highly dynamic nature of RNA, which has previously been shown to impact small-molecule binding,23,24 and based on our initial hit criteria.
Two-Dimensional Screen to Identify Ligands Selective for pre-miR-21
To identify compounds with selectivity for pre-miR-21, we measured CRCs for each hit for inhibition of Dicer-mediated cleavage of pre-miR-21, our assay target, and pre-let-7d, another pre-miRNA substrate. Of note, CRCs were not obtained for the NPEs since they are mixtures; however, triplicate confirmation analysis was performed against both pre-miR-21 and pre-let-7d to enable two-dimensional screening. Major differences between these pre-miRNA substrates include the number of internal loops present within the double-stranded region and the sizes of the terminal loops (Fig. 3). These structural features have previously been shown to be important in small-molecule binding.25 Moreover, they have also been found to impact Dicer kinetics;26,27 however, we hypothesized that this would not be problematic, as our assay is performed under single-turnover conditions to ensure sufficient S/B for HTS, and the majority of pre-miRNAs have been shown to be efficiently cleaved under similar conditions.27
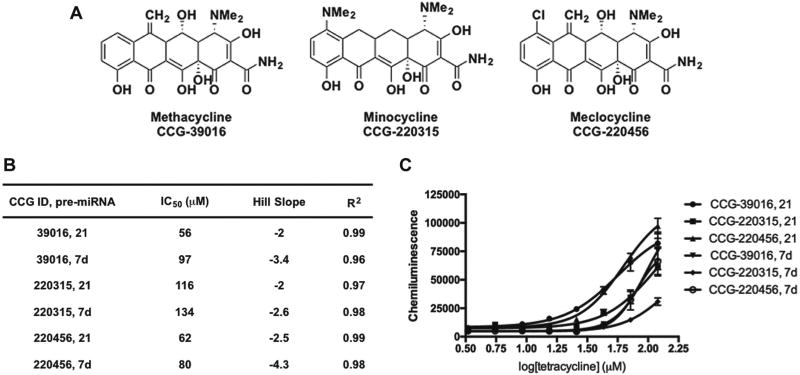
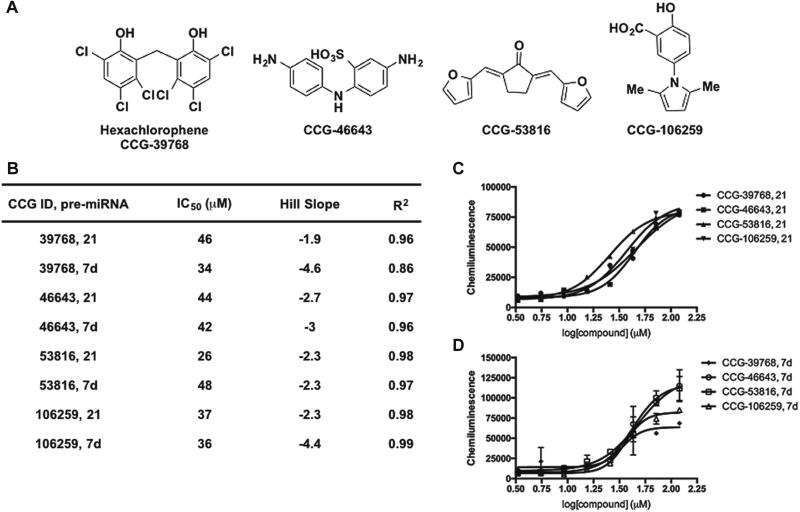
The structures of potent dose-responsive hit compounds identified from the screen are shown in Figure 4A; unfortunately, none of these scaffolds were found to exhibit selectivity for pre-miR-21 (Figs. 4 and 5). With respect to known chemical space for targeting RNA, the 30S ribosome-binding tetracyclines,28 methacycline, minocycline, and meclocycline (Fig. 4B), emerged as hits. Although tetracyclines are established RNA ligands, this antibiotic class has been avoided in rational design strategies, likely due to synthetic difficulties,29 chemical instability,30 and weaker binding affinity over other promiscuous scaffolds, like the aminoglycosides.31 Interestingly, we have previously shown that the aminoglycosides, streptomycin7 and kanamycin (data not shown), do not inhibit Dicer-mediated pre-miR-21 maturation. This finding was also observed by researchers who carried out a fluorescence-based assay of Dicer cleavage of a long, double-stranded RNA substrate.32 Thus, tetracyclines may be a better scaffold for the future rational design of potentially selective pre-miRNA binders. Of the remaining small-molecule hits (Fig. 5B), most are known promiscuous scaffolds22 that contain potentially reactive groups, which likely contributes to their mechanism of action, whether it be RNA binding or nonspecific inhibition of Dicer.
Figure 4.
Confirmation of tetracycline hits from cat-ELCCA for pre-miRNA maturation. (A) Structures. (B) Tabulated data from CRCs. (C) CRCs from 3.3 to 120 µM; data are represented as the mean ± SD from duplicate analysis.
Figure 5.
Confirmation of small-molecule hits from cat-ELCCA for pre-miRNA maturation. (A) Structures. (B) Tabulated data from CRCs. (C,D) CRCs from 3.3 to 120 µM for pre-miR-21 and pre-let-7d, respectively; data are represented as the mean ± SD from duplicate analysis.
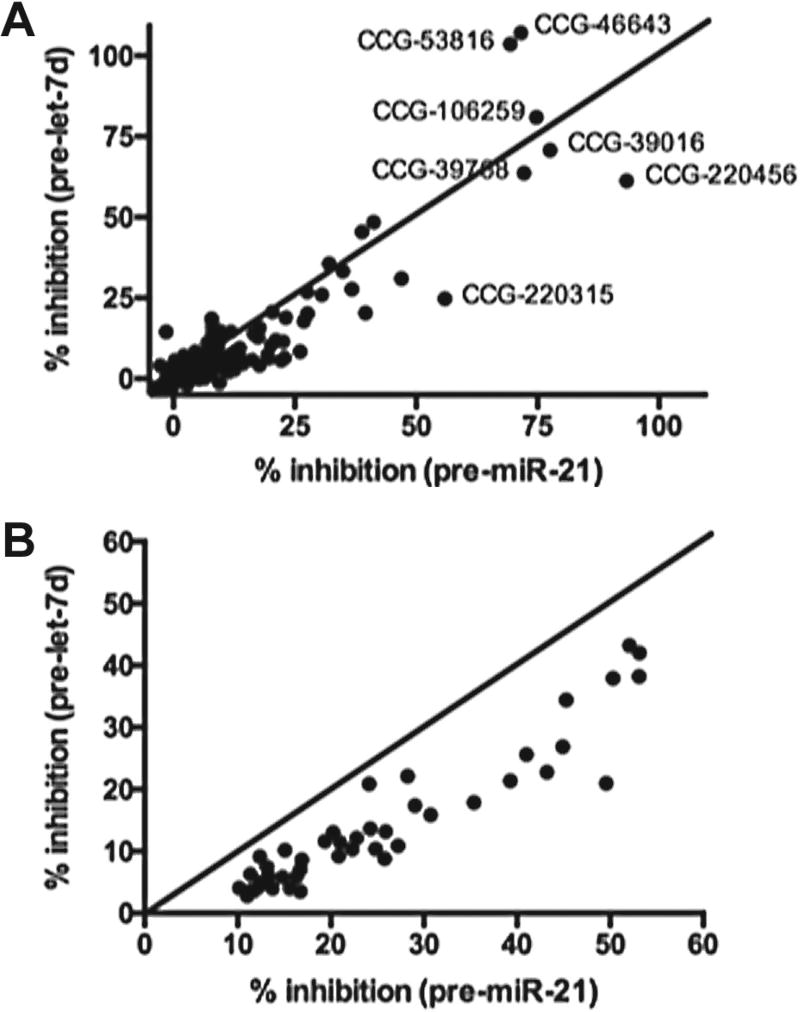
To further analyze our small-molecule and NPE hits and identify any weak binders that may exhibit some selectivity for pre-miR-21, we performed two-dimensional analysis of our screening results. The results of this analysis after plotting the normalized average percent inhibitions for pre-miR-21 versus pre-let-7d processing are shown in Figure 6A,B. From this analysis, it is clear that the majority of small molecules tested fall along the midway line and exhibit little to no selectivity. Selectivity ratios ranged between 0- and 67-fold, with a mean of 1.8-fold and median of 1.05-fold. Compounds exhibiting higher selectivity typically exhibited very weak activity (3%–8% inhibition), so this is likely not significant. On the other hand, for the NPEs, the samples all skewed toward pre-miR-21, with selectivity ratios of 1.1- to 4.8-fold (mean of 2.2-fold and median of 2-fold). We are currently engaged in the follow-up of select NPEs, and future work will determine if natural product scaffolds are capable of exhibiting higher selectivity for an RNA target, especially as purified samples.
Figure 6.
Two-dimensional analysis of average percent inhibition for (A) small-molecule hits (120 µM) and (B) NPE hits (75 µg/mL).
Discussion
The selective targeting of RNA with small molecules remains an unsolved problem in medicinal chemistry.33,34 Most efforts in the area have focused on the engineering of known RNA-binding scaffolds (e.g., aminoglycosides and benzimidazoles) in the hope of attaining selectivity;33,34 however, only in a few cases has this been reported as successful.35,36 With respect to the discovery of new chemical space for targeting RNA, large-scale screening campaigns have been performed on only select RNAs, including HIV TAR and Rev Response Element (RRE) RNAs and the HCV IRES RNA.37–43 High-throughput phenotypic cellular assays of SMN1 mRNA splicing have revealed promising hits that were later developed into clinical candidates,44,45 yet sequence-selective RNA binding has only been demonstrated for one of these scaffolds.45 More recently, a screen of compounds with antibacterial activity led to the discovery of ribocil, a selective small-molecule modulator of bacterial riboflavin riboswitches,46 providing further evidence that noncoding RNAs can be targeted with small molecules.
In this report, we described our efforts toward discovering new chemical space for the selective targeting of pre-miRNAs via HTS of small-molecule and NPE libraries. From this work, two major conclusions can be made; the first regards the assay methodology used, and the second regards library composition for RNA targeting. To enable this line of investigation, we used our group’s newly developed cat-ELCCA for Dicer-mediated pre-miRNA maturation.7,8 As this was the first time that any cat-ELCCA has been used in HTS, we were excited to see that it performed excellently with high reproducibility (Z′ factor of 0.52–0.63), demonstrating its potential for this goal. From these results, we are encouraged to apply cat-ELCCA for HTS against other targets, such as RNA-–protein and protein–protein interactions, due to its many benefits over traditional fluorescence-based biochemical assays.5–8
With respect to library composition, as small molecules, we used the commercial compound libraries available at our institution’s screening center. Although these libraries are most accessible across academia, from our results, it is evident that they are likely not composed of molecules that can bind avidly and specifically to an RNA of interest. On the other hand, from our NPE screening data, expansion to these new areas of chemical space appears to be promising. Thus, we propose that researchers interested in drug discovery for RNA targets expand their screening efforts to these more highly functionalized areas of chemical space, which could also include larger and diverse pharmaceutical industry libraries. Our laboratory is currently involved in applying this strategy toward pre-miR-21, the efforts of which will be disclosed in due course.
Acknowledgments
The authors would like to thank Atsunori Kaneshige for assistance with NPE screening and Renju Jacob for assistance in the preparation of Figure 2B.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported through a pilot grant from the University of Michigan Center for the Discovery of New Medicines and the NIH (R01 GM118329 to A.L.G. and P30 CA046592 to the University of Michigan Comprehensive Cancer Center).
Footnotes
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Ha M, Kim VN. Regulation of microRNA Biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Rana TM. Therapeutic Targeting of microRNAs: Current Status and Future Challenges. Nat. Rev. Drug Disc. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 3.Rupaimoole R, Slack FJ. microRNA Therapeutics: Towards a New Era for the Management of Cancer and Other Diseases. Nat. Rev. Drug Disc. 2017;16:203–221. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 4.White PJ, Anastasopoulos F, Pouton CW, et al. Overcoming Biological Barriers to In Vivo Efficacy of Antisense Oligonucleotides. Expert Rev. Mol. Med. 2009;11:e10. doi: 10.1017/S1462399409001021. [DOI] [PubMed] [Google Scholar]
- 5.Garner AL, Janda KD. cat-ELCCA: A Robust Method to Monitor the Fatty Acid Acyltransferase Activity of Ghrelin O-Acyltransferase (GOAT) Angew. Chem. Int. Ed. 2010;49:9630–9634. doi: 10.1002/anie.201003387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garner AL, Janda KD. A Small Molecule Antagonist of Ghrelin O-Acyltransferase (GOAT) Chem. Commun. 2011;47:7512–7514. doi: 10.1039/c1cc11817j. [DOI] [PubMed] [Google Scholar]
- 7.Lorenz DA, Song JM, Garner AL. High-Throughput Platform Assay Technology for the Discovery of Pre-microRNA-Selective Small Molecule Probes. Bioconj. Chem. 2015;26:19–23. doi: 10.1021/bc500544v. [DOI] [PubMed] [Google Scholar]
- 8.Lorenz DA, Garner AL. A Click Chemistry-Based microRNA Maturation Assay Optimized for High-Throughput Screening. Chem. Commun. 2016:8267–8270. doi: 10.1039/c6cc02894b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz PG, Auld DS, Schultz PJ, et al. Titration-Based Screening for Evaluation of Natural Product Extracts: Identification of an Aspulvinone Family of Luciferase Inhibitors. Chem. Biol. 2011;18:1442–1452. doi: 10.1016/j.chembiol.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J-H, Chung TDY, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 11.Jacob RT, Larsen MJ, Larsen SD, et al. MScreen: An Integrated Compound Management and High-Throughput Screening Data Storage and Analysis System. J. Biomol. Screen. 2012;17:1080–1087. doi: 10.1177/1087057112450186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volinia S, Calin GA, Liu C-G, et al. A microRNA Expression Signature of Human Solid Tumors Defines Cancer Gene Targets. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medina PP, Nolde M, Slack FJ. OncomiR Addiction in an In Vivo Model of microRNA-21-Induced Pre-B-Cell Lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 14.Selcuklu SD, Donoghue MTA, Spillane C. miR-21 as a Key Regulator of Oncogenic Processes. Biochem. Soc. Trans. 2009;37:918–925. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- 15.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a Role in Cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 16.Chirayil S, Chirayil R, Luebke KJ. Discovering Ligands for a microRNA Precursor with Peptoid Microarrays. Nucleic Acids Res. 2009;37:5486–5497. doi: 10.1093/nar/gkp549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz JP, Chirayil R, Chirayil S, et al. Association of a Peptoid Ligand with the Apical Loop of pri-miR-21 Inhibits Cleavage by Drosha. RNA. 2014;20:528–539. doi: 10.1261/rna.042911.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Yang F, Zubovic L, et al. Targeting Inhibition of Oncogenic miR-21 Maturation with Designed RNA-Binding Proteins. Nat. Chem. Biol. 2016;12:717–723. doi: 10.1038/nchembio.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connelly CM, Boer RE, Moon MH, et al. Discovery of Inhibitors of microRNA-21 Processing Using Small Molecule Microarrays. ACS Chem. Biol. 2017;12:435–443. doi: 10.1021/acschembio.6b00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGovern SL, Caselli E, Grigorieff N, et al. A Common Mechanism Underlying Promiscuous Inhibitors from Virtual and High-Throughput Screening. J. Med. Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 21.McGovern SL, Helfand BT, Feng B, et al. A Specific Mechanism of Nonspecific Inhibition. J. Med. Chem. 2003;46:4265–4272. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 22.Baell JB, Holloway GA. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 23.Getz M, Sun X, Casiano-Negroni A, et al. NMR Studies of RNA Dynamics and Structural Plasticity Using NMR Residual Dipolar Couplings. Biopolymers. 2007;86:384–402. doi: 10.1002/bip.20765. [DOI] [PubMed] [Google Scholar]
- 24.Stelzer AC, Frank AT, Kratz JD, et al. Discovery of Selective Bioactive Small Molecules by Targeting an RNA Dynamic Ensemble. Nat. Chem. Biol. 2011;7:553–559. doi: 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tran T, Disney MD. Identifying the Preferred RNA Motifs and Chemotypes That Interact by Probing Millions of Combinations. Nat. Commun. 2012;3:1125. doi: 10.1038/ncomms2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakravarthy S, Sternberg SH, Kellenberger CA, et al. Substrate-Specific Kinetics of Dicer-Catalyzed RNA Processing. J. Mol. Biol. 2010;404:392–402. doi: 10.1016/j.jmb.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Y, Zhang X, Graves P, et al. A Comprehensive Analysis of Precursor microRNA Cleavage by Human Dicer. RNA. 2012;18:2083–2092. doi: 10.1261/rna.033688.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brodersen DE, Clemons WM, Jr, Carter AP, et al. The Structural Basis for the Action of the Antibiotics Tetracycline, Pactamycin, and Hygromycin B on the 30S Ribosomal Subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 29.Liu F, Myers AG. Development of a Platform for the Discovery and Practical Synthesis of New Tetracycline Antibiotics. Curr. Opin. Chem. Biol. 2016;32:48–57. doi: 10.1016/j.cbpa.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Leeson LJ, Weidenheimer JF. Stability of Tetracycline and Riboflavin. J. Pharm. Sci. 1969;58:355–357. doi: 10.1002/jps.2600580315. [DOI] [PubMed] [Google Scholar]
- 31.Aboul-ela F. Strategies for the Design of RNA-Binding Small Molecules. Future Med. Chem. 2010;2:93–119. doi: 10.4155/fmc.09.149. [DOI] [PubMed] [Google Scholar]
- 32.Podolska K, Sedlak D, Bartunek P, et al. Fluorescence-Based High-Throughput Screening of Dicer Cleavage Activity. J. Biomol. Screen. 2014;19:417–426. doi: 10.1177/1087057113497400. [DOI] [PubMed] [Google Scholar]
- 33.Thomas JR, Hergenrother PJ. Targeting RNA with Small Molecules. Chem. Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 34.Guan L, Disney MD. Recent Advances in Developing Small Molecules Targeting RNA. ACS Chem. Biol. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- 35.Disney MD, Angelbello AJ. Rational Design of Small Molecules Targeting Oncogenic Noncoding RNAs from Sequence. Acc. Chem. Res. 2016;49:2698–2704. doi: 10.1021/acs.accounts.6b00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rzuczek SG, Colgan LA, Nakai Y, et al. Precise Small-Molecule Recognition of a Toxic CUG RNA Repeat Expansion. Nat. Chem. Biol. 2017;13:188–193. doi: 10.1038/nchembio.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mei H-Y, Mack DP, Galan AA, et al. Discovery of Selective, Small-Molecule Inhibitors of RNA Complexes. I. The Tat Protein/TAR RNA Complexes Required for HIV-1 Transcription. Bioorg. Med. Chem. 1997;5:1173–1184. doi: 10.1016/s0968-0896(97)00064-3. [DOI] [PubMed] [Google Scholar]
- 38.Mei H-Y, Cui M, Lemrow SM, et al. Discovery of Selective, Small-Molecule Inhibitors of RNA Complexes. II. Self-Splicing Group I Intron Ribozyme. Bioorg. Med. Chem. 1997;5:1185–1195. doi: 10.1016/s0968-0896(97)00065-5. [DOI] [PubMed] [Google Scholar]
- 39.Chapman RL, Stanley TB, Hazen R, et al. Small Molecule Modulators of HIV Rev/Rev Response Element Interaction Identified by Random Screening. Antiviral Res. 2002;54:149–162. doi: 10.1016/s0166-3542(01)00222-4. [DOI] [PubMed] [Google Scholar]
- 40.Gooding KB, Higgs R, Hodge B, et al. High-throughput screening of library compounds against an oligonucleotide substructure of an RNA target. J. Am. Soc. Mass Spectrom. 2004;15:884–892. doi: 10.1016/j.jasms.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Seth PP, Miyaji A, Jefferson EA, et al. SAR by MS: Discovery of a New Class of RNA-Binding Small Molecules for the Hepatitis C Virus: Internal Ribosome Entry Site IIA Subdomain. J. Med. Chem. 2005;48:7099–7102. doi: 10.1021/jm050815o. [DOI] [PubMed] [Google Scholar]
- 42.Baugh C, Wang S, Li B, et al. SCAN—A High-Throughput Assay for Detecting Small Molecule Binding to RNA Targets. J. Biomol. Screen. 2009;14:219–229. doi: 10.1177/1087057108330111. [DOI] [PubMed] [Google Scholar]
- 43.Warui DM, Baranger AM. Identification of Small Molecule Inhibitors of the HIV-1 Nucleocapsid-Stem-Loop 3 RNA Complex. J. Med. Chem. 2012;55:4132–4141. doi: 10.1021/jm2007694. [DOI] [PubMed] [Google Scholar]
- 44.Naryshkin NA, Weetall M, Dakka A, et al. Motor Neuron Disease. SMN2 Splicing Modifiers Improve Motor Function and Longevity in Mice with Spinal Muscular Atrophy. Science. 2014;345:688–693. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- 45.Palacino J, Swalley SE, Song C, et al. SMN2 Splice Modulators Enhance U1-pre-mRNA Association and Rescue SMA Mice. Nat. Chem. Biol. 2015;11:511–517. doi: 10.1038/nchembio.1837. [DOI] [PubMed] [Google Scholar]
- 46.Howe JA, Wang H, Fischmann TO, et al. Selective Small-Molecule Inhibition of an RNA Structural Element. Nature. 2015;526:672–677. doi: 10.1038/nature15542. [DOI] [PubMed] [Google Scholar]