Abstract
Chronic inflammation has a crucial role in cancer development and the progression of various tumors, including pancreatic ductal adenocarcinoma (PDAC). The arachidonate cascade is a major inflammatory pathway that produces several metabolites, such as prostaglandin E2. The enzyme 15‐hydroxyprostaglandin dehydrogenase (15‐PGDH) degrades prostaglandin and is frequently decreased in several types of cancer; however, the molecular mechanisms of 15‐PGDH suppression are unclear. The current study was carried out to elucidate the molecular mechanisms and clinical significance of 15‐PGDH suppression in PDAC. Here, we showed that interleukin‐1β (IL‐1β), a pro‐inflammatory cytokine, downregulates 15‐PGDH expression in PDAC cells, and that IL‐1β expression was inversely correlated with 15‐PGDH levels in frozen PDAC tissues. We also found that activated macrophages produced IL‐1β and reduced 15‐PGDH expression in PDAC cells. Furthermore, the number of CD163‐positive tumor‐associated macrophages was shown to be inversely correlated with 15‐PGDH levels in PDAC cells by immunohistochemical staining of 107 PDAC samples. Finally, we found that low 15‐PGDH expression was significantly associated with advanced tumors, presence of lymph node metastasis and nerve invasion, and poor prognosis in PDAC patients. Our results indicate that IL‐1β derived from TAMs suppresses 15‐PGDH expression in PDAC cells, resulting in poor prognosis of PDAC patients.
Keywords: 15‐Hydroxyprostaglandin dehydrogenase, interleukin‐1β, pancreatic ductal adenocarcinoma, tumor microenvironment, tumor‐associated macrophage
Abbreviations
- 15‐PGDH
15‐hydroxyprostaglandin dehydrogenase
- CM
conditioned medium
- IHC
immunohistochemistry
- IL‐1β
interleukin‐1β
- LPS
lipopolysaccharide
- PDAC
pancreatic ductal adenocarcinoma
- PGE2
prostaglandin E2
- TAM
tumor‐associated macrophage
- TNF‐α
tumor necrosis factor‐α
1. INTRODUCTION
Pancreatic ductal adenocarcinoma is one of the most lethal and aggressive cancers because it is frequently diagnosed at an advanced stage, when this cancer is ineligible for complete surgical resection and is resistant to therapy.1 Furthermore, recurrence is common even after complete resection and is usually only amenable to palliative chemotherapy. The principal risk factor of PDAC is chronic pancreatitis, which results in an approximately 13‐fold increased risk for PDAC.2
Chronic inflammation is implicated in the development of several cancers, including liver, stomach, colon, uterus, bladder, and pancreatic cancers. The relationships between hepatitis virus and hepatocellular carcinoma, Helicobacter pylori and gastric cancer, and human papilloma virus and cervical cancer have been confirmed by overwhelming evidence.3, 4, 5, 6, 7 Therefore, anti‐inflammatory agents such as aspirin, a COX inhibitor that blocks prostaglandin synthases, have been reported to reduce the risk of many cancers.8, 9
Solid tumors are composed of cancer cells and diverse types of stromal cells, including cancer‐associated fibroblasts and TAMs.10, 11, 12, 13 Thus, tumor progression depends on not only the aggressive characteristics of cancer cells themselves but also on their interactions with stromal cells. A previous study reported that prostaglandin signal activation caused by TAMs promoted tumor metastasis.14 In addition, prostaglandin signaling had an important role in gastric tumorigenesis through TAM recruitment.15, 16 Given these reports, prostaglandin signaling in the arachidonate cascade likely has an impact on cancer progression in inflammatory environments.
Recently, 15‐PGDH, which degrades prostaglandin, has attracted attention, and inhibition of 15‐PGDH caused PGE2 accumulation and promoted tissue regeneration in mice by expanding the tissue stem cell fraction.17 Although 15‐PGDH is known to be a tumor suppressor in colon, lung, and breast cancers,18, 19, 20 the relationship between 15‐PGDH expression and PDAC progression is still unclear. Here, we elucidated the mechanism underlying regulation of 15‐PGDH expression and the prognostic impact of 15‐PGDH expression in PDAC.
2. MATERIALS AND METHODS
2.1. Patients and tissue samples
Primary PDAC tissues were obtained from 127 PDAC patients who underwent radical pancreatic resection at Kumamoto University Hospital (Kumamoto, Japan) between April 2002 and December 2015. Signed informed consent to participate was obtained from all patients. The study was approved by the Medical Ethics Committee of Kumamoto University (IRB approval no. 1291).
2.2. Cell lines, macrophages, and cell culture
The human PDAC cell lines PK‐8 and S2‐013 were obtained from the Japanese Collection of Research Bioresource Cell Bank (Ibaraki, Japan) and RIKEN Bioresource Center Cell Bank (Tsukuba, Japan). The cell lines were cultured in RPMI‐1640 supplemented with 10% FBS. Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. Cells were tested and confirmed negative for mycoplasma before use. Macrophages were obtained from PBMCs as previously described.21 For the direct and indirect co‐culture assays, the cell lines and macrophages were cultured in RPMI‐1640.
2.3. Preparation of CM
We prepared CM from PK‐8 cells and macrophages as previously described.22 Briefly, PK‐8 cells and macrophages were seeded (1 × 105 cells/mL) into 100‐mm dishes. After 72 hours, the culture medium was transferred to centrifuge tubes and centrifuged at 3000 g for 5 minutes at room temperature. The supernatant was used as CM without being frozen.
2.4. Recombinant proteins, antibodies, and siRNA transfection
Recombinant human IL‐1β and TNF‐α (Tonbo Biosciences, San Diego, CA, USA) were purchased. Antibodies against the following proteins were used as primary antibodies: β‐actin (#4967; Cell Signaling Technology), CD163 (10D6; Leica Biosystems, Wetzlar, Germany), and 15‐PGDH (ab187161; Abcam).
Silencer Select siRNAs were purchased from Thermo Fisher Scientific (Rockford, IL, USA); the siRNA IDs of si‐15‐PGDH #1 and #2 are s6879 and s6880, respectively. Cultured cells were transfected with the siRNAs (final concentration, 5 μM using Lipofectamine RNAiMAX Reagent; Thermo Fisher Scientific). Cell viability was measured using Trypan blue and calculated as the number of viable cells divided by the total number of cells.
2.5. Immunohistochemistry and scoring
Paraffin‐embedded sections obtained from the PDAC patients were deparaffinized and soaked in distilled water. Sample processing and IHC procedures were performed as described below. Endogenous peroxidase activity was blocked using 3% hydrogen peroxide. Sections were incubated with diluted primary antibodies. Detection was undertaken with a biotin‐free HRP enzyme‐labeled polymer of the Envision Plus detection system (Dako, Tokyo, Japan). A positive reaction was visualized using diaminobenzidine solution followed by counterstaining with Mayer's hematoxylin. All IHC staining was scored blind by two investigators independently. Immunohistochemical staining of 15‐PGDH was scored based on both the intensity and the extent of cell staining. The average proportion of positively stained cells was estimated and given a percentage score on a scale from 1 to 6: 1, 0%‐5%; 2, 6%‐20%; 3, 21%‐40%; 4, 41%‐60%; 5, 61%‐80%; and 6, 81%‐100%. The average intensity of positively stained cells was given an intensity score from 0 to 3: 0, absent; 1, weak; 2, moderate; and 3, strong expression). The two scores were multiplied to characterize 15‐PGDH expression as low (0‐7) or high (8‐18). CD163‐positive cells were counted in four randomly selected area of a high‐power field (×200) of a microscope. Immunohistochemical staining of CD163 was scored as the average absolute number of stained cells in a high‐power field.
2.6. Whole‐cell lysate extraction and Western blot analysis
Cultured cells were washed twice with cold PBS and lysed in RIPA buffer containing 1 × Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). The lysate was sonicated for 1 minute at 1‐second intervals and centrifuged at 15 000 g for 15 minutes at 4°C. The supernatant was collected as a whole‐cell lysate.
Protein samples were subjected to SDS‐PAGE and transferred to PVDF membranes in Towbin buffer. The membranes were blocked with 5% low‐fat, dry milk in TBST (25 mM Tris‐HCl [pH 7.4], 125 mM NaCl, and 0.1% Tween‐20). The membrane was incubated with primary antibodies in Can Get Signal Solution 1 (Toyobo, Osaka, Japan) at 4°C overnight. The signals were detected after incubation with rabbit or mouse secondary antibodies in Can Get Signal 2 at room temperature for 1 hour, using an ECL Detection System (GE Healthcare, Little Chalfont, UK).
2.7. Real‐time RT‐PCR
Total RNA was extracted from cultured cells using a miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Complementary DNA was synthesized from the total RNA in the presence of SuperScript III, RNaseOUT Recombinant Ribonuclease Inhibitor, Random Primer, and Oligo(dT)12‐18 Primer (Thermo Fisher Scientific). Quantitative RT‐PCR was carried out using the TaqMan probe method. All primers and probe sets were designed by the Universal Probe Library Assay Design Center (Basel, Switzerland). All primers were synthesized by Genenet, and all probes were purchased from Roche Diagnostics (Basel, Switzerland).
The quantitative RT‐PCR assay was carried out in the presence of 1 × LightCycler 480 Probes Master (Roche Diagnostics), 0.1 μM Universal Probe Library Probes, 0.5 μM each forward and reverse primers, and cDNA in a total volume of 10 μL. The reaction was performed using a LightCycler 480 System II (Roche Diagnostics). All reactions were carried out in triplicate according to the manufacturer's recommended thermocycling conditions. Relative expression values for each gene were calculated using the 2−ΔΔCT method with normalization to β‐actin. The primer sequences and probes used in real‐time PCR were as follows: 15‐PGDH (HPGD_#13), 5′‐CCAAAGACATTGATAAAGCCATAA‐3′ and 5′‐CACGCCATAGCAATTCACC‐3′; β‐actin (ACTB_#11), 5′‐ATTGGCAATGAGCGGTTC‐3′ and 5′‐CGTGGATGCCACAGGACT‐3′; IL‐1β (IL1B_#10), 5′‐AAAGCTTGGTGATGTCTGGTC‐3′ and 5′‐GGACATGGAGAACACCACTTG‐3′; and CD163 (CD163_#50), 5′‐GAAGATGCTGGCGTGACAT‐3′ and 5′‐GCTGCCTCCACCTCTAAGTC‐3′.
2.8. Enzyme‐linked immunosorbent assay
Protein levels of IL‐1ß were measured by ELISA (R&D Systems) in conditioned medium. The assay procedure was performed according to the manufacturer's protocol. Protein values were calculated as pg/mL.
2.9. Flow cytometry
The protocol for analyzing CD68, CD163, and IL‐1β by flow cytometry has been described in a previous study.23 Briefly, the cells were stained using Fluorescein isothiocyanate (FITC)‐conjugated anti‐CD68 antibody (BioLegend, San Diego, CA, USA), phycoerythrin (PE)‐conjugated anti‐CD163 antibody (BioLegend), and allophycocyanin (APC)‐conjugated anti‐IL‐1β antibody (BioLegend). Isotype‐matched control antibodies were also obtained from BioLegend. Flow cytometry analysis was undertaken with a FACSVerse flow cytometer (BD, Franklin Lakes, NJ, USA).
2.10. Statistical analysis
All experiments were carried out in triplicate, and the data shown are representative of consistently observed results. The data are presented as the mean ± SE. The Mann‐Whitney U test was used to compare continuous variables between the two groups. Categorical variables were compared using a χ2‐test. Survival curves were constructed using the Kaplan‐Meier method, and the log‐rank test was used to evaluate the statistical significance of differences. For statistical analyses, we used JMP (version 9; SAS Institute, Cary, NC, USA), taking into consideration the assumptions required for the respective tests. P‐values less than .05 were considered statistically significant. The investigators were blinded to allocation for IHC analyses and the in vitro experiments.
3. RESULTS
3.1. Low 15‐PGDH expression is implicated in poor PDAC prognosis
To examine the impact of 15‐PGDH expression on the prognosis of PDAC patients, we first examined 15‐PGDH expression in cancer cells using IHC analysis of PDAC patients who underwent radical pancreatic resection between 2002 and 2015. Among 127 PDAC patients, we excluded 14 patients who underwent R1 surgery, because R status is one of the strongest prognostic factors,24 and 6 patients who had less than 12 months of follow‐up after surgery because of an inability to perform an accurate survival analysis. Thus, we undertook the IHC analysis using primary tissues obtained from 107 PDAC patients (Figure 1A). Expression of 15‐PGDH was detected in the cytoplasm in PDAC cells (Figure 1B). A high level of 15‐PGDH was observed in 47 patients. We further assessed the relationship between 15‐PGDH expression and clinicopathological factors of PDAC patients. Low 15‐PGDH expression was significantly associated with advanced tumor stage (P = .004), presence of lymph node metastasis (P = .002), and presence of nerve invasion (P = .013) (Table 1). A Kaplan‐Meier survival analysis revealed that low 15‐PGDH expression was significantly associated with poor prognosis of relapse‐free survival (P = .0464; Figure 1C) and overall survival (P = .0461; Figure 1D). These results suggested that low 15‐PGDH expression in PDAC cells is correlated with tumor progression and poor prognosis in PDAC patients.
Figure 1.
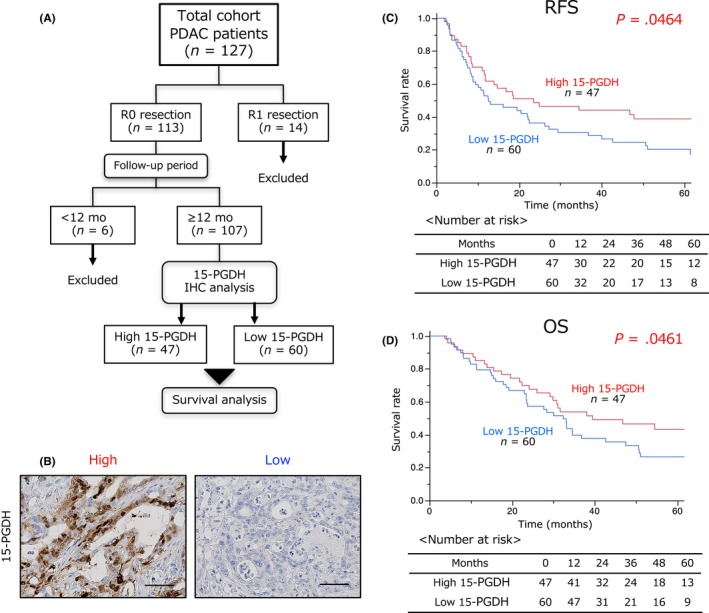
Low 15‐hydroxyprostaglandin dehydrogenase (15‐PGDH) expression is implicated in poor pancreatic ductal adenocarcinoma (PDAC) prognosis. A, Workflow diagram of patients who underwent pancreatic resection and contributed samples for immunohistochemical (IHC) analysis. B, Representative IHC staining of 15‐PGDH expression in 107 PDAC tissues. Scale bar = 100 μm. C,D, Relationship between 15‐PGDH expression and relapse‐free survival (C) or overall survival (D) using the Kaplan‐Meier method
Table 1.
Relationship between 15‐hydroxyprostaglandin dehydrogenase (15‐PGDH) expression and clinicopathological findings in 107 patients with pancreatic ductal adenocarcinoma
| 15‐PGDH expression | |||
|---|---|---|---|
| High | Low | ||
| Clinicopathological factor | n = 47 | n = 60 | P‐value |
| Age, years, mean (range) | 69 (46‐82) | 69 (43‐90) | .806 |
| Sex, male / female | 26/21 | 26/34 | .218 |
| CEA, ng/mL | 1.7 (0.2‐39.3) | 2.2 (0.4‐112) | .643 |
| CA19‐9, U/L | 36 (0.1‐4760) | 64.8 (0.1‐3722) | .874 |
| T stage, 1‐2 / 3‐4 | 15/32 | 6/54 | .004 |
| Tumor size, mm | 29 (5‐60) | 30 (10‐50) | .340 |
| LN metastasis, positive / negative | 20/27 | 43/17 | .002 |
| Nerve invasion, positive / negative | 36/11 | 56/4 | .013 |
| Ly invasion, positive / negative | 26/21 | 42/18 | .118 |
| V invasion, positive / negative | 36/11 | 42/18 | .444 |
CA19‐9, carbohydrate antigen 19‐9; CEA, carcinoembryonic antigen; LN, lymph node; Ly, lymphatic; V, venous.
3.2. Downregulation of 15‐PGDH by IL‐1β leads to PDAC cell growth
To investigate whether the expression level of 15‐PGDH affects tumor growth in PDAC cells, we silenced 15‐PGDH expression using two siRNAs. HPGD (the gene coding 15‐PGDH protein) expression and 15‐PGDH protein level were significantly reduced by both siRNAs in two types of PDAC cells (Figure 2A,B). In addition, the silencing of 15‐PGDH significantly increased cell growth in the two types of PDAC cells (Figure 2C,D). These results indicate that 15‐PGDH suppresses cell growth and functions as a tumor suppressor in PDAC cells.
Figure 2.
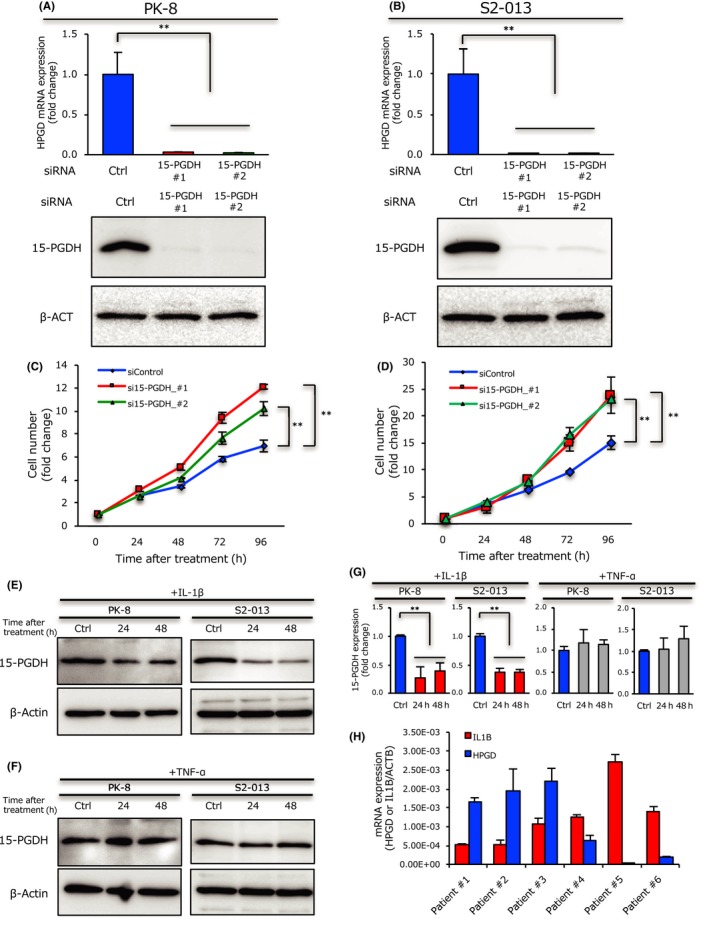
15‐Hydroxyprostaglandin dehydrogenase (15‐PGDH) downregulation by interleukin‐1β (IL‐1β) enhances pancreatic ductal adenocarcinoma cell growth. A,B, Expression of HPGD (the gene coding 15‐PGDH protein, upper panel) or 15‐PGDH (lower panel) in PK‐8 cells (A) or S2‐013 cells (B) after treatment with siRNA targeting 15‐PGDH or with control siRNA, evaluated by quantitative RT‐PCR (upper panel) or Western blot analysis (lower panel). Data are presented as the treated/control cell ratio. C,D, PK‐8 cells (C) or S2‐013 cells (D) transfected with siRNAs targeting 15‐PGDH or with control siRNA were incubated for up to 96 hours and assayed for cell number; data are presented as the treated/control (time = 0) cell ratio. E,F, Expression of 15‐PGDH in PK‐8 cells or S2‐013 cells after IL‐1β (E) or tumor necrosis factor‐α (TNF‐α) (F) treatment for 24 and 48 hours and distilled water treatment for 48 hours as a control was evaluated by Western blotting. G, Column graph showing relative 15‐PGDH levels in PK‐8 cells or S2‐013 cells after IL‐1β and TNF‐α treatment for 24 and 48 hours, and distilled water treatment for 48 hours as a control, were evaluated using ImageJ software. H, Expression of HPGD and IL1B in six PDAC patients determined by quantitative RT‐PCR. Data were normalized to the ACTB mRNA level and are shown as the mean ± SD of three independent experiments. **P < .01
Recent studies have indicated that TNF‐α suppresses 15‐PGDH expression in colorectal cancer25 and that IL‐1β also suppresses 15‐PGDH expression in gastric cancer.26 Thus, we examined the effect of these cytokines on 15‐PGDH expression in PDAC cells. Consequently, we found that IL‐1β treatment significantly suppressed 15‐PGDH expression in the two types of PDAC cells; however, TNF‐α treatment had no effect on 15‐PGDH expression in PDAC cells (Figure 2E‐G). We next assessed the relationship between IL‐1β and 15‐PGDH expression in frozen PDAC tumor tissues from six patients with resectable PDAC. Interestingly, IL‐1β expression was inversely correlated with 15‐PGDH expression (Figure 2H). These results indicate that IL‐1β suppresses HPGD expression in PDAC cells in frozen tissues as well as in PDAC cell lines.
3.3. Activated macrophages produce IL‐1β and decrease 15‐PGDH expression in PDAC cells
A previous study showed that macrophages have a central role in inflammation, and activated macrophages produce many types of pro‐inflammatory cytokines following exposure to Toll‐like receptor agonists such as LPS.27 We therefore hypothesized that activated macrophages produce IL‐1β in the tumor microenvironment, and the concentration of IL‐1β in the conditioned medium of PDAC cells and macrophages was examined by ELISA. Notably, we found that LPS‐induced activated macrophages produced abundant IL‐1β compared with PDAC cells or non‐activated macrophages (Figure 3A). We further investigated whether activated macrophages can alter 15‐PGDH expression in PDAC cells using indirect co‐culture assays. Western blot analysis revealed that only activated macrophages reduced 15‐PGDH expression in PDAC cells, and non‐activated macrophages did not show these changes (Figure 3B). We also found that CM obtained from activated macrophages reduced 15‐PGDH expression in PDAC cells, although CM from PK‐8 and non‐activated macrophages did not induce such changes (Figure 3C). Tumor‐associated macrophages have been implicated in tumor progression, and CD163 was identified as a potent marker of TAMs.28 Therefore, we confirmed that LPS‐induced activated macrophages significantly increased CD163 expression compared with non‐activated macrophages (Figure 3D). Moreover, we also found that macrophages directly co‐cultured with PDAC cells had higher CD163 expression than macrophages without co‐culture (Figure 3E). We next assessed whether TAMs altered 15‐PGDH expression through IL‐1β secretion under direct co‐culture conditions. The PDAC cells co‐cultured with non‐activated and activated macrophages had lower 15‐PGDH expression levels than PDAC cells alone (Figure 3F,G). We further assessed whether IL‐1β was predominantly expressed in TAMs under direct co‐culture conditions using flow cytometry analysis. As a result, we observed that, under direct co‐culture conditions, IL‐1β is more highly expressed in TAMs than in PK‐8 cells (Figure 3H,I,S1). These results suggest that activated macrophages secrete abundant IL‐1β and suppress 15‐PGDH expression in PDAC cells.
Figure 3.
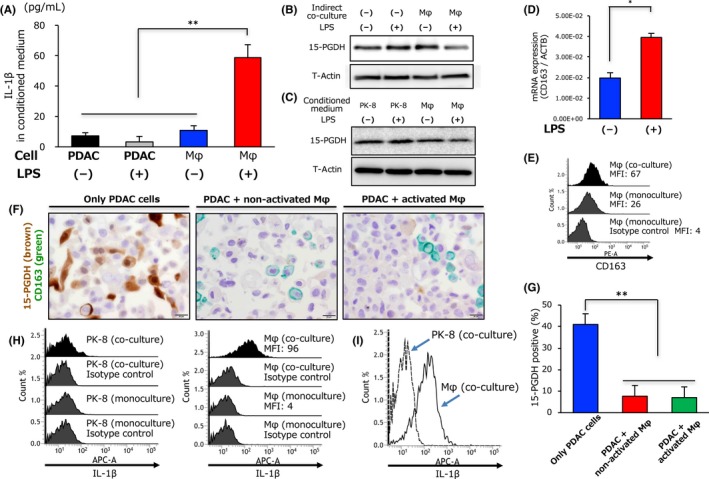
Activated tumor‐associated macrophages produce interleukin‐1β (IL‐1β) and reduce 15‐hydroxyprostaglandin dehydrogenase (15‐PGDH) expression in pancreatic ductal adenocarcinoma cells. A, Concentration of IL‐1β in conditioned medium from PK‐8 cells or macrophages (Mφ) treated with lipopolysaccharide (LPS) or distilled water (DW) was evaluated by ELISA. B, Expression of 15‐PGDH in PK‐8 cells determined by Western blot analysis 72 hours after co‐culture with or without macrophages treated with LPS or DW. C, Expression of 15‐PGDH in PK‐8 cells determined by Western blot analysis 72 hours after treatment with conditioned medium from PK‐8 or macrophages treated with LPS or DW. D, Quantitative RT‐PCR analysis of CD163 mRNA expression in macrophages 24 hours after LPS or DW treatment; data are presented as the LPS‐treated / control expression ratio. E, Expression levels of CD163 in monocultured macrophages (middle panel) or macrophages co‐cultured with PK‐8 cells (upper panel) were evaluated by flow cytometry. F, Representative double‐immunohistochemical staining of 15‐PGDH (brown) and CD163 (green) in only PK‐8 cells (left panel), PK‐8 cells directly co‐cultured with non‐activated macrophages (middle panel), or PK‐8 cells directly co‐cultured with activated macrophages (right panel). Scale bar = 20 μm. G, Column graph showing the percentage of 15‐PGDH‐positive cells in only PK‐8 cells, PK‐8 cells directly co‐cultured with non‐activated macrophages, or PK‐8 cells directly co‐cultured with activated macrophages. H,I, Expression levels of IL‐1β in monocultured PK‐8 cells or PK‐8 cells co‐cultured with macrophages and in macrophages co‐cultured with PK‐8 cells were evaluated by flow cytometry. **P < .01. APC‐A, allophycocyanin; MFI, mean fluorescence intensity; PE‐A, phycoerythrin
3.4. Number of infiltrating TAMs is inversely correlated with 15‐PGDH expression in PDAC cells
Given that TAMs secrete IL‐1β and suppress 15‐PGDH expression in PDAC cells in vitro, we further investigated the relationship between 15‐PGDH expression in PDAC cells and the number of infiltrating TAMs in human PDAC tissues. We examined CD163 expression using IHC analysis to detect infiltrating TAMs in 107 human PDAC samples (Figure 4A,B), and the median number of TAMs was 20 cells/high‐power field (range, 2‐42 cells/high‐powered field). Consequently, the number of infiltrating TAMs was found to be inversely correlated with 15‐PGDH expression, in agreement with the direct co‐culture assay results (Figure 4C). These findings suggest that infiltrating TAMs in PDAC tissues suppress 15‐PGDH expression in PDAC cells.
Figure 4.
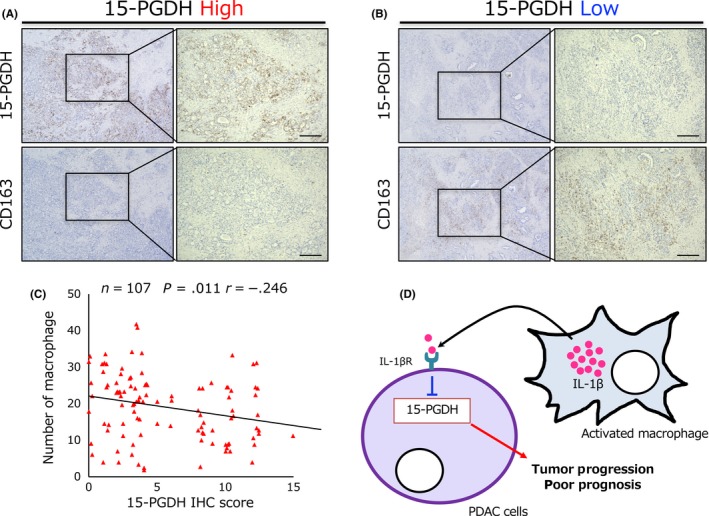
Tumor‐associated macrophages are inversely correlated with pancreatic ductal adenocarcinoma (PDAC) cells harboring high 15‐hydroxyprostaglandin dehydrogenase (15‐PGDH) expression. A,B, Representative immunohistochemical (IHC) staining of 15‐PGDH (upper panel) and CD163 (lower panel) expression in high 15‐PGDH (A) and low 15‐PGDH (B) serial PDAC specimens. Scale bar = 200 μm. C, Graph showing Pearson's correlation between the expression of 15‐PGDH and the number of CD163‐positive cells in 107 PDAC patients. D, Schematic representation of the findings of this study. IL‐1βR, interleukin‐1β receptor
4. DISCUSSION
Cancer‐related inflammation has been shown to promote tumor growth and progression in diverse cancers, including PDAC.29 The arachidonate cascade, a major inflammatory pathway, produces several metabolites due to activation of COX, and tumors harboring high COX expression have been associated with poor prognosis of patients with various types of cancer.30 Furthermore, 15‐PGDH is the key enzyme responsible for degradation and biological inactivation of PGE2; thus, decreased 15‐PGDH expression leads to PGE2 accumulation in the cells. According to previous studies, HPGD has been reported to be a tumor suppressor gene in several types of cancer.18, 19, 20 Here, we showed that 15‐PGDH expression in PDAC cells is suppressed due to IL‐1β derived from TAMs, and low expression of 15‐PGDH was significantly associated with poor prognosis along with increased tumor progression in PDAC (Figure 4D). Although 15‐PGDH expression is regulated by IL‐1β and TNF‐α, based on our results, only IL‐1β reduced 15‐PGDH expression in PDAC cells.
Tumor‐associated macrophages have been implicated in promoting tumor progression and poor prognosis in many cancers.10 Although various mechanisms are known to enhance tumor escape from the immune system, angiogenesis for tumor progression, and intravasation for tumor invasion, the mechanisms underlying downregulation of the tumor suppressor gene HPGD in cancer have not been elucidated. Notably, CD163 is a potent marker of M2‐polarized macrophages, and CD163‐positive TAMs were implicated in poor prognosis in several cancers.21, 31, 32 In fact, LPS‐treated activated macrophages significantly increased CD163 expression. Furthermore, we also showed that the number of CD163‐positive TAMs is inversely associated with low 15‐PGDH expression in PDAC cells from human PDAC tissues. However, further investigations are needed to establish the functional differences between M1‐ and M2‐polarized macrophages in PDAC.
In summary, we revealed a possible mechanism of promoting cancer progression through inflammatory cytokines in the tumor microenvironment. Given that 15‐PGDH expression in PDAC cells is suppressed due to IL‐1β derived from TAMs and is significantly associated with poor prognosis, the inhibition of this interaction between PDAC cells and TAMs could be a potential strategy for PDAC treatment.
CONFLICT OF INTEREST
The authors have no conflict of interest.
Supporting information
ACKNOWLEDGMENTS
This work was supported in part by the Japan Society for the Promotion of Science (KAKENHI Grant Nos. 16H06257, 16K15595, and 17K16570).
Arima K, Komohara Y, Bu L, et al. Downregulation of 15‐hydroxyprostaglandin dehydrogenase by interleukin‐1β from activated macrophages leads to poor prognosis in pancreatic cancer. Cancer Sci. 2018;109:462–470. https://doi.org/10.1111/cas.13467
Contributor Information
Hideo Baba, Email: hdobaba@kumamoto-u.ac.jp.
Takatsugu Ishimoto, Email: taka1516@kumamoto-u.ac.jp.
REFERENCES
- 1. Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:2140‐2141. [DOI] [PubMed] [Google Scholar]
- 2. Raimondi S, Lowenfels AB, Morselli‐Labate AM, Maisonneuve P, Pezzilli R. Pancreatic cancer in chronic pancreatitis; aetiology, incidence, and early detection. Best Pract Res Clin Gastroenterol. 2010;24:349‐358. [DOI] [PubMed] [Google Scholar]
- 3. zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342‐350. [DOI] [PubMed] [Google Scholar]
- 4. Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4:688‐694. [DOI] [PubMed] [Google Scholar]
- 5. Fukase K, Kato M, Kikuchi S, et al. Effect of eradication of Helicobacter pylori on incidence of metachronous gastric carcinoma after endoscopic resection of early gastric cancer: an open‐label, randomised controlled trial. Lancet. 2008;372:392‐397. [DOI] [PubMed] [Google Scholar]
- 6. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245‐1255. [DOI] [PubMed] [Google Scholar]
- 7. Ishimoto T, Izumi D, Watanabe M, et al. Chronic inflammation with Helicobacter pylori infection is implicated in CD44 overexpression through miR‐328 suppression in the gastric mucosa. J Gastroenterol. 2015;50:751‐757. [DOI] [PubMed] [Google Scholar]
- 8. Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX‐2. N Engl J Med. 2007;356:2131‐2142. [DOI] [PubMed] [Google Scholar]
- 9. Rothwell PM, Fowkes FG, Belch JF, Ogawa H, Warlow CP, Meade TW. Effect of daily aspirin on long‐term risk of death due to cancer: analysis of individual patient data from randomised trials. Lancet. 2011;377:31‐41. [DOI] [PubMed] [Google Scholar]
- 10. Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263‐266. [DOI] [PubMed] [Google Scholar]
- 11. Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239‐252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sugihara H, Ishimoto T, Watanabe M, et al. Identification of miR‐30e* regulation of Bmi1 expression mediated by tumor‐associated macrophages in gastrointestinal cancer. PLoS ONE. 2013;8:e81839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ishimoto T, Miyake K, Nandi T, et al. Activation of transforming growth factor beta 1 signaling in gastric cancer‐associated fibroblasts increases their motility, via expression of rhomboid 5 homolog 2, and ability to induce invasiveness of gastric cancer cells. Gastroenterology. 2017;153:191‐204. e16 [DOI] [PubMed] [Google Scholar]
- 14. Majumder M, Xin X, Liu L, Girish GV, Lala PK. Prostaglandin E2 receptor EP4 as the common target on cancer cells and macrophages to abolish angiogenesis, lymphangiogenesis, metastasis, and stem‐like cell functions. Cancer Sci. 2014;105:1142‐1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ishimoto T, Oshima H, Oshima M, et al. CD44 + slow‐cycling tumor cell expansion is triggered by cooperative actions of Wnt and prostaglandin E2 in gastric tumorigenesis. Cancer Sci. 2010;101:673‐678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oshima H, Hioki K, Popivanova BK, et al. Prostaglandin E(2) signaling and bacterial infection recruit tumor‐promoting macrophages to mouse gastric tumors. Gastroenterology. 2011;140:596‐607. e7 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y, Desai A, Yang SY, et al. TISSUE REGENERATION. Inhibition of the prostaglandin‐degrading enzyme 15‐PGDH potentiates tissue regeneration. Science. 2015;348:aaa2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wolf I, O'Kelly J, Rubinek T, et al. 15‐hydroxyprostaglandin dehydrogenase is a tumor suppressor of human breast cancer. Cancer Res. 2006;66:7818‐7823. [DOI] [PubMed] [Google Scholar]
- 19. Huang G, Eisenberg R, Yan M, et al. 15‐Hydroxyprostaglandin dehydrogenase is a target of hepatocyte nuclear factor 3beta and a tumor suppressor in lung cancer. Cancer Res. 2008;68:5040‐5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mehdawi LM, Prasad CP, Ehrnstrom R, Andersson T, Sjolander A. Non‐canonical WNT5A signaling up‐regulates the expression of the tumor suppressor 15‐PGDH and induces differentiation of colon cancer cells. Mol Oncol. 2016;10:1415‐1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Komohara Y, Ohnishi K, Kuratsu J, Takeya M. Possible involvement of the M2 anti‐inflammatory macrophage phenotype in growth of human gliomas. J Pathol. 2008;216:15‐24. [DOI] [PubMed] [Google Scholar]
- 22. Izumi D, Ishimoto T, Miyake K, et al. CXCL12/CXCR4 activation by cancer‐associated fibroblasts promotes integrin beta1 clustering and invasiveness in gastric cancer. Int J Cancer. 2016;138:1207‐1219. [DOI] [PubMed] [Google Scholar]
- 23. Horlad H, Ma C, Yano H, et al. An IL‐27/Stat3 axis induces expression of programmed cell death 1 ligands (PD‐L1/2) on infiltrating macrophages in lymphoma. Cancer Sci. 2016;107:1696‐1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Neoptolemos JP, Stocken DD, Dunn JA, et al. Influence of resection margins on survival for patients with pancreatic cancer treated by adjuvant chemoradiation and/or chemotherapy in the ESPAC‐1 randomized controlled trial. Ann Surg. 2001;234:758‐768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Otani T, Yamaguchi K, Scherl E, et al. Levels of NAD(+)‐dependent 15‐hydroxyprostaglandin dehydrogenase are reduced in inflammatory bowel disease: evidence for involvement of TNF‐alpha. Am J Physiol Gastrointest Liver Physiol. 2006;290:G361‐G368. [DOI] [PubMed] [Google Scholar]
- 26. Thiel A, Ganesan A, Mrena J, et al. 15‐hydroxyprostaglandin dehydrogenase is down‐regulated in gastric cancer. Clin Cancer Res. 2009;15:4572‐4580. [DOI] [PubMed] [Google Scholar]
- 27. Iliodromiti Z, Anastasiadis A, Varras M, et al. Monocyte function in the fetus and the preterm neonate: immaturity combined with functional impairment. Mediators Inflamm. 2013;2013:753752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Komohara Y, Hirahara J, Horikawa T, et al. AM‐3K, an anti‐macrophage antibody, recognizes CD163, a molecule associated with an anti‐inflammatory macrophage phenotype. J Histochem Cytochem. 2006;54:763‐771. [DOI] [PubMed] [Google Scholar]
- 29. Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860‐867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10:181‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hasita H, Komohara Y, Okabe H, et al. Significance of alternatively activated macrophages in patients with intrahepatic cholangiocarcinoma. Cancer Sci. 2010;101:1913‐1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kridel R, Xerri L, Gelas‐Dore B, et al. The prognostic impact of CD163‐positive macrophages in follicular lymphoma: a study from the BC cancer agency and the lymphoma study association. Clin Cancer Res. 2015;21:3428‐3435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


