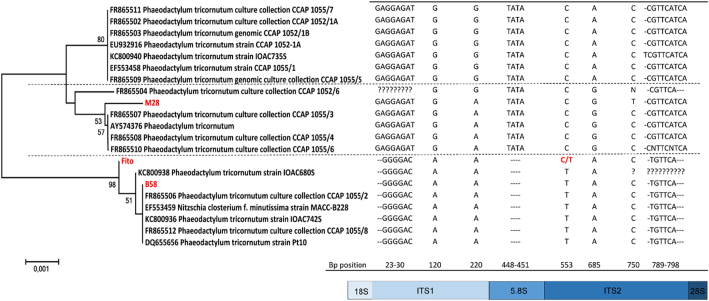
Fig. 3.
Minimal evolution tree displaying the evolutionary relationships in the ITS1-5.8S-ITS2 region of the genome of the three Phaeodactylum tricornutum strains (Fito, M28 and B58) used in the experiment and closest related strains obtained from Genbank. The evolutionary history was inferred using the Minimum Evolution method [22]. The optimal tree with the sum of branch length = 0,01 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [23]. The evolutionary distances were computed using the Maximum Composite Likelihood method [24] and are in the units of the number of base substitutions per site. Evolutionary analyses were conducted in MEGA6 [20].