Abstract
Acinetobacter baumannii are Gram-negative bacilli that pose a constant threat to susceptible patients because of increased resistance to multiple antibiotics and persistence in the hospital environment. After genome analysis, we discovered that A. baumannii harbors genes that share homology to an enzymatic pathway that elongates long-chain fatty acids (LCFA) in fungi. Previously, 1,2,4-triazolidine-3-thiones (T-3-Ts) were shown to inhibit hyphae production in fungi, and this same LCFA elongation pathway was implicated as the possible target. Therefore, we investigated if T-3-Ts also have activity against multidrug-resistant A. baumannii. Surprisingly, all of the clinical isolates of A. baumannii that were tested have susceptibility to ECC145 and ECC188 with MIC90 values of 8.0 µg/mL. In contrast, reference strains and clinical isolates of other common nosocomial bacteria that lack the LCFA pathway also lacked susceptibility. Time–kill experiments revealed that both ECC145 and ECC188 have a bacteriostatic effect against A. baumannii. Mass spectrometry analysis suggested that exposure to T-3-Ts resulted in less LCFA production. Supplementation of media with either 0.02% w/v oleic or linoleic acid abrogated the bacteriostatic effect of the compounds, which again implicated LCFA elongation as the target. Our results suggest these molecules could be a promising start to further exploit what appears to be an important aspect of A. baumannii membrane function and integrity.
Keywords: ESKAPE pathogens, fatty acids, antibiotic, antibacterial, fungi, Gram-negative membrane
Graphical abstract
To date, limited progress has been made in developing new antibacterial treatments for multidrug-resistant (MDR) and extremely drug-resistant (XDR) ESKAPE pathogens, despite the need for new treatments.1,2 The acronym ESKAPE, which stands for Enterococcus faecium (E), Staphylococcus aureus (S), Klebsiella pneumoniae (K), Acinetobacter baumannii (A), Pseudomonas aeruginosa (P), Enterobacter species (E), is reflective of these bacterial species that have the ability to “escape” antibiotic treatment in clinical settings.3 Of these species, perhaps the most troubling are the Gram-negative bacterial species K. pneumoniae and A. baumannii because of their rapid acquisition of resistance to antibiotic treatments4 and because the mortality rates in susceptible patients can be high, sometimes >50%.5,6 A. baumannii is particularly worrying as it has been shown that this species can also serve as a reservoir for antibiotic resistance genes for other bacterial species.7
The Dunn laboratory at the Uniformed Services University of the Health Sciences has spent many years evaluating the enzymes required for long-chain fatty acid (LCFA) synthesis and elongation in a variety of organisms. In 2009, it was discovered that Tsc10p from Saccharomyces cerevisiae had a mammalian homologue, FVT1, which could complement Tsc10p function in yeast, and that this protein is the 3-ketosphinganine reductase for mammalian cells.8 In addition to this work, the homology of Tsc10p to prokaryotic genomes was also evaluated, with homologues discovered in multiple bacterial taxa and species; however, it was noted that, of these, A. baumannii was one of the only common nosocomial pathogens to have a homologue of this gene. The acquisition of fungal DNA by A. baumannii is a possible evolutionary event given that the A. baumannii genome is very plastic,9–11 and these bacteria likely interact with12,13 and potentially kill fungi14 in the human body.
As genome sequencing has become more affordable and relatively easy to execute, multiple genomes of A. baumannii have now become available for analysis.10,11,15,16 Therefore, we evaluated the A. baumannii genomes for genes that might be related to LCFA synthesis and elongation using BLAST and BLASTp.17 Specifically, we discovered that many genes in the Candida albicans and S. cerevisiae LCFA biosynthetic pathway had homologues to genes in the AB5075 genome,18 as well as an Ole1p homologue [(expected value = 4.0 × 10−51, identities = 100/239 (42%), positives = 147/239 (62%)], which is annotated as a Δ9 stearoyl-CoA desaturase responsible for converting saturated fatty acyl-CoA substrates to monounsaturated fatty acids (GenBank: AKA30381.1).19,20 Therefore, we performed a larger BLASTp analysis against all Acinetobacter species and found that the Ole1p homologue was present in all sequenced A. baumannii strains as well as other Acinetobacter species linked to infection including A. lwoffii, A. junii, A. nosocomialis, and A. ursingii.
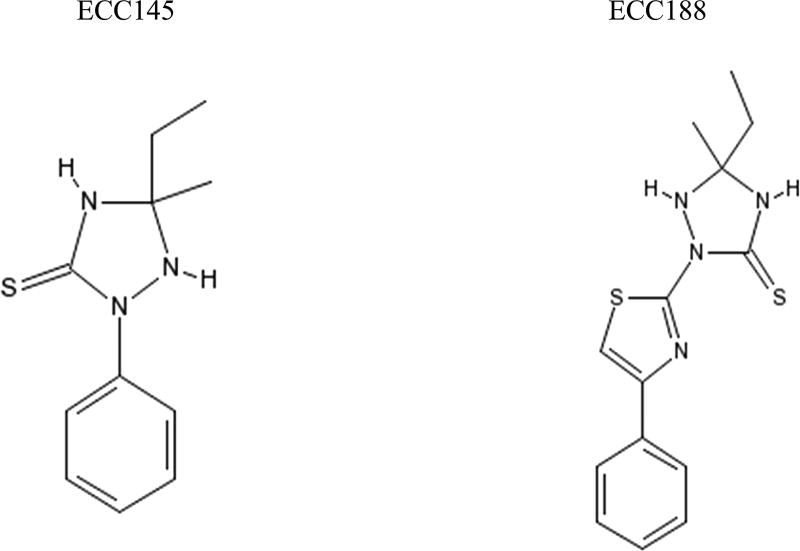
The Ole1p homologue was specifically of interest because a previous study by Xu et al. had described a series of ChemBridge library compounds, 1,2,4-Triazolidine-3-thiones (T-3-Ts), that had whole-cell antifungal activity, and they had speculated that the target of these compounds was the gene OLE1, which makes the Ole1p protein.21 Specifically, the OLE1± C. albicans strain had hypersensitivity to two of the T-3-Ts, ECC145 and ECC188 (Figure 1), which were shown to block hypha formation in C. albicans, a known mechanism of virulence.21 Given that A. baumannii possessed an Ole1p homologue, we decided to explore if the same compounds examined by Xu et al., ECC145 and ECC188, had antibacterial activity against A. baumannii as well.
Figure 1.
Chemical structures of the 1,2,4-triazolidine-3-thiones, ECC145 and ECC188, used in this study.
RESULTS AND DISCUSSION
Susceptibility
Both ECC145 and ECC188 were evaluated against type strains of ESKAPE pathogens and clinical isolates to include S. aureus, a Gram-positive pathogen, and Escherichia coli, which are also linked to hospital-acquired infections and have a relatively different set of genes with respect to metabolism and physiology when compared to Acinetobacter.22 Only the A. baumannii strains showed susceptibility at concentrations below 128 µg/mL for either ECC145 or ECC188 (Table 1). Both compounds had activity against A. baumannii with MIC50 and MIC90 values of 8.0 and 16 µg/mL, respectively. In contrast, other ESKAPE pathogens had MIC values >128 µg/mL, and an MIC of 512 µg/mL was not uncommon (Table 1).
Table 1.
MICa of ECC145 and ECC188 against Common Nosocomial Bacterial Pathogens
| MIC for ECC145 (µg/mL) | MIC for ECC188 (µg/mL) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| organism (n) | 50% | 90% | range | 50% | 90% | range |
| Escherichia coli (10) | >512 | >512 | >512 | >512 | >512 | 256–>512 |
| Staphylococcus aureus (9) | 256 | 256 | 128–>512 | 256 | 512 | 128–>512 |
| Klebsiella pneumoniae (10) | >512 | >512 | >512 | >512 | >512 | >512 |
| Acinetobacter baumannii (44) | 8 | 8 | 2–16 | 4 | 8 | 1–8 |
| Pseudomonas aeruginosa (10) | 512 | 512 | 128–>512 | 512 | 512 | 256–>512 |
MIC in cation-adjusted Mueller–Hinton broth, 22 h.
To further evaluate the activity of ECC145 and ECC188, we evaluated a diverse set of 44 different A. baumannii isolates, where the diversity of 33 of these isolates has been previously detailed.23 For both compounds, the MIC90 was 8.0 µg/mL for these isolates (Table 1). The range of MIC varied from 2.0 to 16 µg/mL for ECC145, and the range of MIC for ECC188 was between 1.0 and 8.0 µg/mL (Table 1). These results suggest that the target of ECC145 and ECC188 is shared among a multitude of different A. baumannii strains. Importantly, no strain, despite the diversity, displayed an inherent resistance to these compounds when using this assay.
Time–Kill Assay
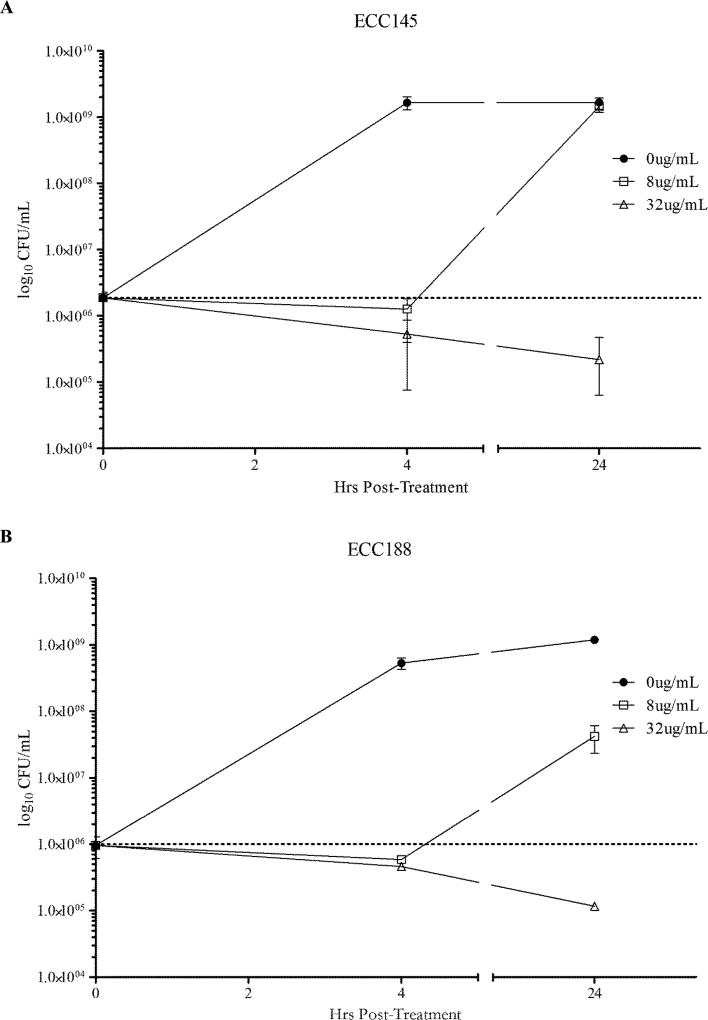
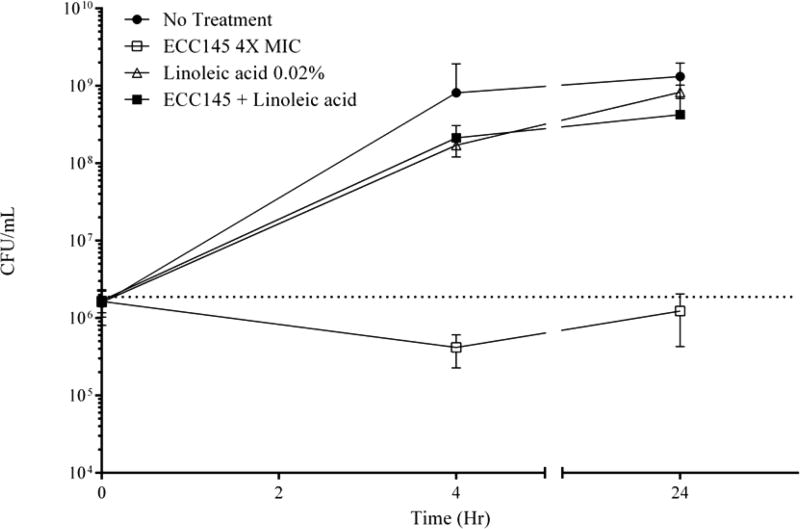
The observed inhibition was further characterized by the time–kill assay. AB5075, a reference strain,23 was challenged with 1 or 4 times the determined MIC for either ECC145 or ECC188 (Figure 2). Cultures treated with ECC145 at 1 times the MIC showed no growth at 4 h with a mean CFU of 1.27 × 106 CFU/mL, but grew to untreated levels of 1.45 × 109 CFU/mL by 24 h, whereas cultures treated with 4 times the MIC had CFU/mL reduced to 5.33 × 105 at 4 h post-treatment and were further reduced to 2.19 × 105 by 24 h post-treatment (Figure 2A). Cultures treated with ECC188 at 1 times the MIC showed a similar initial bacteriostatic effect at 4 h, but reached a mean CFU/mL of 4.22 × 107 after 24 h (Figure 2B). Similarly, cultures treated with 4 times the MIC of ECC188 had CFU/mL reduced to 5.87 × 105 by 4 h and were reduced nearly 1.0 log to 1.17 × 105 CFU/mL at 24 h post-treatment. These results suggest the mechanism of action of T-3-Ts against A. baumannii is bacteriostatic.
Figure 2.
Time–kill assays displaying mean and standard error of the mean of CFU/mL at time of initial treatment and at 4 and 24 h post-treatment for ECC145 (A) and ECC188 (B) at 1 and 4 times the MIC determined via microbroth dilution.
Transmission Electron Microscopy Analysis
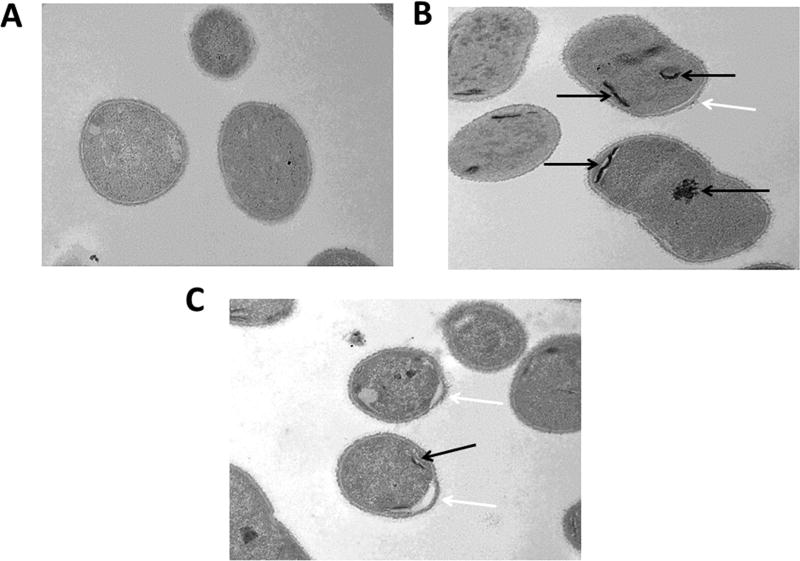
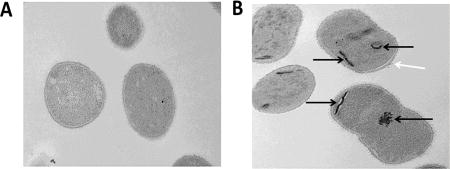
To further understand the mechanism of action, we hypothesized that electron microscopy may be able to provide some insight. A. baumannii cells were treated with 8.0 µg/mL ECC145 or 8.0 µg/mL ECC188 for 1 h and were then compared to untreated controls utilizing thin-section transmission electron microscopy. Interestingly, the treated cells appeared to have aggregates or inclusion bodies (Figure 3B,C), whereas untreated bacteria did not harbor these darker areas of staining (Figure 3A). Also, the membrane in treated cells appeared to be disrupted, especially at the poles, where a portion of membrane extended from the rest of the bacterium (Figure 3B,C). Again, this phenotype was not observed in untreated cells (Figure 3A).
Figure 3.
Thin-section transmission electron microscopy images (20,000×) of AB5075 untreated (A), exposed to ECC145 at 8.0 µg/mL (B), or exposed to ECC188 at 8.0 µg/mL (C) for 1 h. Aggregates are visible in the cytoplasm (black arrows), and membrane disruption (white arrows) is visible on the bacterial surface, distended from the cytoplasm, of the treated bacteria.
Mass Spectrometry Analysis
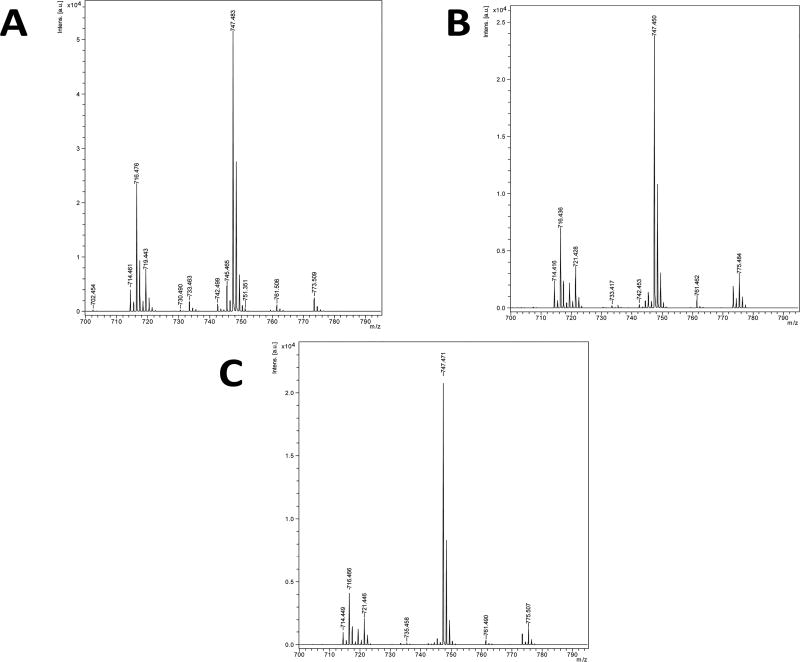
On the basis of the EM data, we speculated that the negative impact on growth could be linked to overall deficiencies in the membrane, similar to what was seen in fungi when treated with ECC145 and ECC188.21 To further evaluate the effects of T-3-Ts treatment on A. baumannii, we analyzed the overall fatty acid content of the membrane of the bacteria via mass spectrometry in the negative ion mode. AB5075 was exposed to ECC145 or ECC188 for 6 h at 1 times the MIC (8.0 µg/mL) or 4 times the MIC (32 µg/mL) and compared to untreated controls. The untreated group showed two major populations of phospholipids, a species at m/z 716.5, phosphatidylethanolamine (PE), and a species at m/z 747.5, phosphatidylglycerol (PG). Both of these species contain 16/18:1 fatty acids attached to the head groups. Treatment with ECC145 and ECC188 over time not only appeared to result in the incorporation of less 18:1 fatty acids to the membrane but also seemed to alter the overall amount of total PE in the samples (Figure 4). Interestingly, the samples treated at 4 times the MIC for 6 h show a large increase in the short-chain PG species indicated at m/z 721.5. Additionally, there was a shift from m/z 773.5 and 747.4 to the species at m/z 775.5 and 749.5, respectively (data not shown). Both of these shifts indicate a change in fatty acid usage from saturated to unsaturated fatty acid and suggest treatment by T-3-Ts abrogates 16/18:1 fatty acid development as was also shown in C. albicans21 and is a known function of Ole1p.19,20
Figure 4.
Mass spectrometry analysis of A. baumannii that was untreated (A) or treated with ECC145 for 6 h (B) or with ECC188 for 6 h (C).
Fatty Acid Supplementation
Because the mass spectrometry and EM data implicated T-3-Ts in the disruption of fatty acid elongation with detrimental consequences to the bacteria, we hypothesized that perhaps A. baumannii could acquire LCFA from the environment to abrogate the effects of the T-3-Ts. To test this hypothesis in vitro, we supplemented CAMHB media used in the MIC assay with 0.02% linoleic acid. The addition of linoleic acid increased the observed MIC of ECC145 for A. baumannii 5075 from 8.0 to >256 µg/mL. This effect was confirmed in a time–kill assay (Figure 5). Whereas cultures treated with ECC145 at 4 times the MIC displayed the previously observed bacteriostatic activity, the cells treated with ECC145 with 0.02% linoleic acid in the CAMHB media showed growth 2 logs above initial inoculum to 2.13 × 108 CFU/mL by 4 h post-treatment and reached a final CFU/mL of 4.26 × 108 after 24 h, closer to that of untreated cultures (1.32 × 109 CFU/mL) (Figure 5). A similar result was seen when A. baumannii was treated with ECC188 (data not shown). Therefore, we tested the addition of other unsaturated fatty acids, specifically oleic acid and Tween 80 (polysorbate 80), which is a complex mixture of polyoxyethylene ethers of partial oleic acid esters of sorbitol anhydrides as well as other intermediates.24 Both of these sources of unsaturated fatty acids also abrogated T-3-T activity in an MIC assay (Table 2), which suggests the target of T-3-Ts is a pathway that involves unsaturated fatty acid and elongation of fatty acids as predicted previously.21
Figure 5.
Time–kill assays displaying the mean and standard error of the mean of CFU/mL at time of initial treatment and at 4 and 24 h post-treatment for AB5075 treated with either ECC145 at 4 times the MIC, linoleic acid at 0.02% w/v, or a combination of ECC145 and linoleic acid.
Table 2.
MIC (µg/mL) of ECC145 and ECC188 against AB5075 ± Unsaturated Fatty Acids
| fatty acid addition | ECC145 | ECC188 |
|---|---|---|
| no addition | 4 | 8 |
| 0.02% linoleic acid | 256 | 256 |
| 0.02% oleic acid | 512 | 512 |
| 0.02% polysorbate 80 | 512 | 512 |
Targeting and Resistance
To determine a target for the T-3-Ts, a number of experiments were attempted. First, we analyzed our transposon mutant libraries of A. baumannii.18,23 The gene encoding the Ole1p is annotated as a stearoyl-CoA 9-desaturase, and the current identifier is ABUW_0611 for the AB5075 genome.18 Given the experimental data from Xu et al. that seemed to implicate Ole1p as the target of T-3-Ts,21 it should be an essential gene in A. baumannii, and any transposon mutant would not have survived when the mutants were first generated. To our surprise, we found multiple transposon mutants in the gene encoding the Ole1p homologue, and ABUW_0611 does not appear to be an essential gene.18 Nonetheless, we tested the ABUW_0611 mutant in the case Ole1p homologue worked in concert with other enzymes to elongate fatty acids, which, in that case, an increase in MIC might have been detected in this mutant background. When the transposon mutants of ABUW_0611 were exposed to ECC145 or ECC188, we did not see any abrogation of activity and the MIC remained 8.0 µg/mL (data not shown). Furthermore, we evaluated the whole pool of transposon mutants in the presence of ECC145 at increasing concentrations of 8.0, 16, and 32 µg/mL to identify mutants that were not susceptible to T-3-Ts. Although some colonies arose on these plates in the first 24 h, none of these bacteria were able to be further passaged, suggesting that if a specific transposon mutant was allowing survival in the presence of ECC145, it was not stable.
In another attempt to determine the target of T-3-Ts, we also checked for the development of spontaneous mutants at 0.5, 1, and 2 times the MIC of ECC145 on plates for 24 h. No colonies arose on any plates over this time period (data not shown). Lastly, we evaluated resistance by serial passaging A. baumannii in the presence of 2-fold increasing concentrations of ECC145 (2–32 µg/mL). After 10 passages, growth was finally observed in the 32 µg/mL tube (data not shown), suggesting a spontaneous mutant occurred that allowed for growth. However, again, we could not further passage from this point into fresh media, suggesting the mutant was not stable.
Further Mechanistic Studies
Given that Ole1p does not appear to be essential in A. baumannii or the target of the compounds, we ran a number of additional experiments to determine whether these compounds could be acting as phenotypic Pan-Assay Interference (PAINS) compounds in our assays. First, given that fatty acid supplementation abrogated activity, we investigated whether this phenomenon was due to simple disruption of ECC145 aggregation. In the presence of 0.01% Triton, we observed no change in MIC, and ECC145 inhibited AB5075 growth at 8 µg/mL. We observed the same activity in the presence of other detergents, such that growth inhibition studies in media supplemented with either 0.1% PEG400 or 0.1% tyloxagol also returned MIC values of 8.0 µg/mL. Thus, aggregation does not appear to play a role in the anti-A. baumannii activity of 3-T-3s.
Next, given the inability to generate stable resistant mutants, we probed whether the mechanism of action could simply be due to membrane disruption. We performed a BacLight assay on AB5075 using 0.5, 1, and 2 times the MIC of ECC145 and observed no significant membrane disruption at any of these concentrations. We also assessed the hemolytic potential of ECC145 and also observed no hemolysis (<3%) up to the highest concentration studied (512 µg/mL). Therefore, membrane disruption does not appear to underpin antimicrobial activity.
Finally, 3-T-3s may have some hydrolytic instability; however, the stability of the lead compound ECC145 has not been established in bacterial growth broth. We performed a stability study in CAMHB and observed steady degradation of ECC145 over a 24 h period (determined by LC analysis). On the basis of these data, we determined the half-life of ECC145 to be ca. 6–8 h under these conditions. Given that degradation is happening on the time scale of the experiment, one additional explanation is that the compounds are not the actual active species, but the degradation products are. To probe this, we allowed ECC145 to incubate in CAMHB for either 4 or 8 h prior to bacterial treatment. The 4 h pre-incubation still delivered a solution that was able to inhibit growth at 8 µg/mL (original concentration); however, the 8 h pre-incubation was inactive, and we observed visible growth at the MIC (8 µg/mL original concentration). Therefore, the degradation products due to hydrolysis of ECC145 do not appear to be the active constituents in this assay.
DISCUSSION
In this study, we found that A. baumannii harbors some genes required for LCFA elongation similar to what is found in yeast and fungi, which could be an attractive target for drug design. Given this bacterial species is rapidly becoming resistant to all clinically used antibiotics, strides to further understand its physiology and what is required for its pathogenesis remain critical. The presence of this LCFA pathway appears to be somewhat unique to A. baumannii when compared to other common nosocomial pathogens, which also speaks to the unique metabolism and biochemistry pathways this bacterial species appears to have encoded in its genome. Whereas P. aeruginosa is often called a “cousin” of Acinetobacter, the genomes share only 58% homology.22 When compared to the E. coli genome, the A. baumannii genome is missing enzymatic pathways, or there are significant changes in pathways, suggesting its somewhat unique metabolism.22
Separately, the versatility and plasticity of the A. baumannii genome was shown given the possible acquisition of these genes from the C. albicans or S. cerevisiae. As stated in the introduction, it is not completely unexpected given the potential interactions between fungi and A. baumannii. This would also not be the first time A. baumannii acquired DNA from other organisms and hijacked the function if it suited the bacteria’s pathogenic lifestyle requirements. For example, A. baumannii utilizes antibiotic resistance genes from various bacterial sources and incorporates this DNA into its chromosome.11 It should also be noted that A. baumannii has a gene that is homologous to Plasmodium species (our unpublished data), and recently it was shown that bacteriophage DNA in its genome encodes small regulatory RNAs that the bacteria have usurped for regulatory functions.25
One drawback to our study is that we do not know if the Ole1p homologue or the other genes we found in the LCFA pathway are expressed and utilized by A. baumannii, and it is certainly possible that other bacterial species, related to Acinetobacter, were a precursor to other more developed genomes of eukaryotes such as fungi. Regardless, at least some of the LCFA genes in the pathway appear functional given the mass spectrometry data that suggest A. baumannii is generating 18:1 and 16:0 fatty acids for incorporation into the membrane (Figure 4). This result is also in agreement with a previous study that detected 18:1 and 16:0 fatty acids as the major lipid components of the A. baumannii membrane via gas chromatography analysis.26 The fact that T-3-Ts can block the production and/or shorten fatty acids indicates that this enzymatic pathway can also be disrupted. However, at this point, the actual target or targets of the compounds are unclear, and all that can be concluded is that the membrane does not contain its typical fatty acid content after treatment with T-3-Ts. This conclusion is further supported by the electron microscopy date that indicates an apparent membrane disruption at the poles and the formation of inclusion bodies in the cytoplasm. Moreover, the supplementation of media with linoleum acid and oleic acid, which upon import by A. baumannii, abrogates the activity of T-3-Ts (Figure 5).
It is hoped that future efforts to find resistant mutants may identify the target of 3-T-3s in A. baumannii. Unexpectedly, the transposon mutant in the Ole1p homologue was still susceptible to T-3-T activity, and this result suggests that the Ole1p homologue is not the target of T-3-Ts. Possibly, T-3-Ts attack multiple enzymes in the pathway, or other desaturases present in the genome could compensate for a loss of function in this mutant background. Another possibility is that T-3-Ts may activate a reciprocal activity rather than block Ole1p function. Recently, a study identified an enzyme in S. cerevisiae, Sct1p, that competes with and antagonizes Ole1p activity.20 Whereas a Sct1p homologue with an E value of <1.0 was not found in the A. baumannii genome (data not shown), it is still possible that there is another protein not detected by BLASTp analysis that could provide an antagonizing function.
Our work is certainly not the first time researchers have tried to exploit fatty acid synthesis as a target for antibacterials. In fact, many groups have developed inhibitors of FabI, the NADH-dependent enoyl reductase from the type II bacterial fatty acid biosynthesis pathway (FAS-II),27 as well as targeting other enzymes in this pathway such as FabF and FabG.28 However, depending on the bacterial species, the addition of exogenous fatty acids can abrogate the activity of LCFA synthesis inhibitors, and fatty acid import in the host can disrupt activity.28,29 Interestingly, in some Gram-negative bacteria, a shunt pathway was also identified to bypass FAS-II.30 When in vivo experiments are attempted in the future, we will need to be mindful of these possibilities in the host environment. Nevertheless, just like the Fab genes, we believe the target of T-3-Ts, once identified, will be important when it comes to possible antibacterial development against A. baumannii.
Given that we have not identified the target of the 3-T-3s, we ran a number of experiments to determine whether the activity was simply due to a PAINS-type mechanism. Aggregation does not appear to drive activity, as ECC145 is equally active in the absence or presence of detergents (Triton, PEG400, or tyloxagol). Generalized membrane disruption also does not appear to drive antibacterial activity, as ECC145 does not significantly disrupt membrane permeation up to 2 times its MIC as determined by a BacLight assay, nor does it have hemolytic activity. The half-life of the compound was determined to be ca. 6–8 h in CAMHB, and activity does not appear to be driven by degradation products as pre-incubation of ECC145 with CAMHB for 8 h abrogated activity. Although this relative hydrolytic instability could be problematic in an in vivo setting, we note that there is ample precedent for hydrolytically unstable antibiotics (i.e., imipenem) that still show tremendous clinical utility.
In conclusion, we demonstrated that T-3-Ts have a pronounced bacteriostatic effect in vitro and that the most likely outcome of this treatment is blocking the production of LCFA required for complete membrane integrity and cell division. We showed that A. baumannii can bypass this antibacterial activity when in the presence of exogenous unsaturated fatty acids added to growth media, which further implicates 18:1 and 16:0 fatty acids being the affected membrane species. With this knowledge, we can try to identify the protein target(s) of T-3-Ts, design compounds with more activity using SAR and ADME analysis for in vivo testing, and begin to think of combinational therapies that synergize with blocking the synthesis and elongation of LCFA.
MATERIALS AND METHODS
Bacterial Strains and Antimicrobial Agents
The bacteria (a total of 83 strains) used in this study included 44 Acinetobacter baumannii, 10 Klebsiella pneumoniae, 10 Escherichia coli, 9 Staphylococcus aureus, and 10 Pseudomonas aeruginosa and can be found in Table 3. All strains were maintained and subcultured in lysogeny broth (LB) in liquid culture or on plates until utilized in various assays outlined below. T-3-Ts, ECC145 and ECC188, were procured from ChemBridge Co. (San Diego, CA, USA). Working stocks were prepared by dissolving the compounds in dimethyl sulfide (DMSO) to a final concentration of 25 mg/mL.
Table 3.
Sources of Bacterial Strains Used in This Study
| bacterial species | straina | sourceb |
|---|---|---|
| Acinetobacter baumannii | AB0057 | Luis Actis |
| Acinetobacter baumannii | 17978 | Luis Actis |
| Acinetobacter baumannii | 19606 | Luis Actis |
| Acinetobacter baumannii | AB307-0294 | Anthony Campagnari |
| Acinetobacter baumannii | A118 | Luis Actis |
| Acinetobacter baumannii | ACICU | Luis Actis |
| Acinetobacter baumannii | AYE | ATCC |
| Acinetobacter baumannii | SDF | ATCC |
| Acinetobacter baumannii | LUH3876 | Luis Actis |
| Acinetobacter baumannii | RUH134 | Luis Actis |
| Acinetobacter baumannii | RUH876 | Luis Actis |
| Acinetobacter baumannii | AB2828 | MRSN |
| Acinetobacter baumannii | AB3340 | MRSN |
| Acinetobacter baumannii | AB3560 | MRSN |
| Acinetobacter baumannii | AB3638 | MRSN |
| Acinetobacter baumannii | AB3785 | MRSN |
| Acinetobacter baumannii | AB3806 | MRSN |
| Acinetobacter baumannii | AB3917 | MRSN |
| Acinetobacter baumannii | AB3927 | MRSN |
| Acinetobacter baumannii | AB4025 | MRSN |
| Acinetobacter baumannii | AB4026 | MRSN |
| Acinetobacter baumannii | AB4027 | MRSN |
| Acinetobacter baumannii | AB4052 | MRSN |
| Acinetobacter baumannii | AB4269 | MRSN |
| Acinetobacter baumannii | AB4448 | MRSN |
| Acinetobacter baumannii | AB4456 | MRSN |
| Acinetobacter baumannii | AB4490 | MRSN |
| Acinetobacter baumannii | AB4498 | MRSN |
| Acinetobacter baumannii | AB4795 | MRSN |
| Acinetobacter baumannii | AB4857 | MRSN |
| Acinetobacter baumannii | AB4878 | MRSN |
| Acinetobacter baumannii | AB4932 | MRSN |
| Acinetobacter baumannii | AB4957 | MRSN |
| Acinetobacter baumannii | AB4991 | MRSN |
| Acinetobacter baumannii | AB5001 | MRSN |
| Acinetobacter baumannii | AB5075 | MRSN |
| Acinetobacter baumannii | AB5197 | MRSN |
| Acinetobacter baumannii | AB5256 | MRSN |
| Acinetobacter baumannii | AB5674 | MRSN |
| Acinetobacter baumannii | AB5711 | MRSN |
| Acinetobacter baumannii | AB3941ColR | Glen Wortmann |
| Acinetobacter baumannii | AB3942ColR | Glen Wortmann |
| Acinetobacter baumannii | AB4106ColR | Glen Wortmann |
| Klebsiella pneumoniae | KP02615 | MRSN |
| Klebsiella pneumoniae | KP02814 | MRSN |
| Klebsiella pneumoniae | KP02843 | MRSN |
| Klebsiella pneumoniae | KP04776 | MRSN |
| Klebsiella pneumoniae | KP04843 | MRSN |
| Klebsiella pneumoniae | KP05540 | MRSN |
| Klebsiella pneumoniae | KP101436 | Clinton Murray/BAMC |
| Klebsiella pneumoniae | KP101488 | Clinton Murray/BAMC |
| Klebsiella pneumoniae | KP101712 | Clinton Murray/BAMC |
| Klebsiella pneumoniae | KP105371 | Clinton Murray/BAMC |
| Pseudomonas aeruginosa | PA4761 | MRSN |
| Pseudomonas aeruginosa | PA4910 | MRSN |
| Pseudomonas aeruginosa | PA4962 | MRSN |
| Pseudomonas aeruginosa | PA5105 | MRSN |
| Pseudomonas aeruginosa | PA5151 | MRSN |
| Pseudomonas aeruginosa | PA105738 | Clinton Murray/BAMC |
| Pseudomonas aeruginosa | PA105777 | Clinton Murray/BAMC |
| Pseudomonas aeruginosa | PA105819 | Clinton Murray/BAMC |
| Pseudomonas aeruginosa | PA105857 | Clinton Murray/BAMC |
| Pseudomonas aeruginosa | PA105880 | Clinton Murray/BAMC |
| Staphylococcus aureus | MRSA5836 | MRSN |
| Staphylococcus aureus | MRSA5971 | MRSN |
| Staphylococcus aureus | MRSA5968 | MRSN |
| Staphylococcus aureus | MRSA6048 | MRSN |
| Staphylococcus aureus | MRSA6081 | MRSN |
| Staphylococcus aureus | MRSA6139 | MRSN |
| Staphylococcus aureus | MRSA103700 | Clinton Murray/BAMC |
| Staphylococcus aureus | MRSA104161 | Clinton Murray/BAMC |
| Staphylococcus aureus | MRSA107261 | Clinton Murray/BAMC |
| Escherichia coli | EC5107 | MRSN |
| Escherichia coli | EC5378 | MRSN |
| Escherichia coli | EC5379 | MRSN |
| Escherichia coli | EC5403 | MRSN |
| Escherichia coli | EC5466 | MRSN |
| Escherichia coli | EC5483 | MRSN |
| Escherichia coli | EC5502 | MRSN |
| Escherichia coli | EC105454 | Clinton Murray/BAMC |
| Escherichia coli | EC105547 | Clinton Murray/BAMC |
| Escherichia coli | EC109497 | Clinton Murray/BAMC |
ColR, colistin-resistant isolates.
MRSN, Multidrug-resistant Organism Repository and Surveillance Network.
Minimum Inhibitory Concentration (MIC)
MIC end points were determined according to the broth microdilution methodology recommended by the Clinical and Laboratory Standards Institute (CLSI). However, one slight modification is utilized; test strains were grown overnight at 37 °C in cation-adjusted Mueller–Hinton broth (CAMHB) and diluted with fresh CAMHB to yield an initial inoculum of approximately 5.0 × 105 CFU/mL in 96-well plates as opposed to using single colonies. Each test strain was challenged with ECC145 or ECC188 across a concentration range from 512 to 1 µg/mL in 2-fold increments. Plates were incubated at 37 °C for 20–22 h before the MIC end points were read. MIC90 and MIC50 are the lowest concentrations of the antibiotic at which 90 and 50% of the isolates were inhibited, respectively. Subsequent MIC analysis included the supplementation of media with either 0.02% w/v oleic, linoleic acid, or polysorbate 80 (Sigma-Aldrich). Additional MIC supplementation assays with detergents were performed with CAMHB as above with 0.01% Triton, 0.1% PEG400, or 0.1% tyloxagol.
Time–Kill Assays
A. baumannii strain 5075 (AB5075) was grown overnight at 37 °C in CAMHB and diluted in 25 mL of fresh CAMHB to yield an initial inoculum of approximately 106 CFU/mL. ECC145 or ECC188 was added to a final concentration of 0, 1, or 4 times the MIC, and cultures were incubated with shaking at 37 °C. Samples were taken at 0, 4, and 24 h post addition of the tested compound. To test the ability of unsaturated fatty acids to abrogate the activity of ECC145, the growth medium was supplemented with 0.2% w/v linoleic acid (Sigma-Aldrich). CFU/mL was determined by dilution and plating utilizing a spiral plater (Advanced Instruments, Inc.) for each sample in triplicate.
Electron Microscopy
AB5075 was grown overnight at 37 °C in CAMHB and diluted in 25 mL of fresh CAMHB to yield an initial inoculum of approximately 106 CFU/mL. ECC145 or ECC188 was added to a final concentration of 0 or 8 µg/mL, and cultures were incubated with shaking at 37 °C. After 1 h, samples were collected from each condition and (fixed using 4% formaldehyde and 1% glutaraldehyde). Samples were rinsed twice in 0.1 M phosphate buffer for 5 min. Samples were then subjected to En-Bloc staining (2% uranyl acetate aqueous) for 30–45 min. Samples were then dehydrated for 15 min in 50, 75, and 95% ethanol (aqueous), followed by two treatments with 100% ethanol, again for 15 min. Samples were then placed in 100% propylene oxide twice for 15 min, followed by 20% Epon and 80% propylene oxide for 60 min, 50% Epon and 50% propylene oxide overnight, and finally 100% Epon for 60 min. The samples were then embedded in preassembled Beem capsules and baked at 70 °C for 12–24 h. Thin sections were prepared on an Ultracut Microtome (Leica). Sections were imaged on a Jeol 1400 electron microscope.
Total Lipid Extraction and Mass Spectrometry Analysis
Total membrane lipids were extracted from whole cells using a two-step Bligh–Dyer extraction protocol. Briefly, to a sample containing 50 mg of dry cells were added 200 µL of water, 250 µL of chloroform, and 500 µL of methanol to generate a monophasic solution. The solution was briefly vortexed and shaken for 3 h at room temperature. Subsequently, to each tube were added 250 µL each of water and chloroform to generate a biphasic solution, and the mixtures were vortexed and shaken for 30 min at room temperature. Phases were separated by centrifugation (3000 rpm for 10 min at room temperature). The lower chloroform phase was removed and evaporated under nitrogen.
Negative ion matrix-assisted laser ionization desorption–time-of-flight tandem mass spectrometry (MALDI-TOF/TOF MS) experiments were performed. Briefly, total lipids were solubilized in 100 µL of chloroform/methanol (2:1, v/v) and spotted (1 µL) directly onto the MALDI sample plate, followed by 1 µL of 100 mg/mL norharman MALDI matrix dissolved in chloroform/methanol/water (3:1.5:0.25, v/v/v). All experiments were performed using a Bruker Autoflex Speed MALDITOF/TOF mass spectrometer (Bruker Daltonics Inc., Billerica, MA, USA). Each spectrum was an average of 500 shots and 50% laser power. ES Tuning Mix (Agilent, Palo Alto, CA, USA) was used as a calibrate standard, calibrated using Agilent Tuning Mix (Agilent Technologies, Foster City, CA, USA).
Hemolysis Assay
Hemolysis assays were performed on mechanically difibrinated sheep blood (Hemostat Laboratories: DSB100). Difibrinated blood (1.5 mL) was placed into a microcentrifuge tube and centrifuged for 10 min at 10,000 rpm. The supernatant was then removed, and then the cells were resuspended in 1 mL of phosphate-buffered saline (PBS). The suspension was centrifuged, the supernatant was removed, and cells were resuspended two additional times. The final cell suspension was then diluted 10-fold. Test compound was added to aliquots of the 10-fold suspension dilution of blood. PBS was used as a negative control and a zero hemolysis marker. Triton × (a 1% sample) was used as a positive control serving as the 100% lysis marker. Samples were then placed in an incubator at 37 °C while being shaken at 200 rpm for 1 h. After 1 h, the samples were centrifuged for 10 min at 10,000 rpm. The resulting supernatant was diluted by a factor of 40 in distilled water. The absorbance of the supernatant was then measured with a UV spectrometer at a 540 nm wavelength.
Bacterial Membrane Permeabilization Assay
The BacLight assay (Invitrogen) was used to assess membrane permeability. AB5075 was grown overnight in CAMHB at 37 °C with shaking. The culture was diluted 1:40 in CAMHB and grown to an optical density at 600 nm (OD600) of ~1.0 (~4 h of growth). The cultures were centrifuged at 10000g for 15 min, the cell pellet was washed once with sterile water, resuspended at 2 times the original volume, and aliquoted, and test compounds were added. Suspensions were incubated at 37 °C with shaking for 1 h and then centrifuged at 10000g for 10 min, washed once with sterile water, and resuspended in water. A 1:1 mixture of SYTO-9 and propidium iodide was added to the suspension (3 µL/mL) and mixed well. One hundred microliters of the suspension was added to each well of a 96-well plate, and the plates were incubated in the dark for 15 min at room temperature. Green fluorescence (SYTO-9) was read at 530 nm, and red fluorescence (propidium iodide) was read at 645 nm (excitation wavelength, 485 nm). The ratio of green to red fluorescence was expressed as a percentage of the control.
Acknowledgments
We kindly acknowledge Teresa Dunn for helpful discussions. We thank the MRSN, Luis Actis, Tony Campagnari, and Glenn Wortmann from Walter Reed National Military Medical Center (WRNMMC) and Katrin Mende and Clint Murray from Brooke Army Medical Center (BAMC) for providing the clinical isolates and the reference strains for our work. We acknowledge the support of the Military Infectious Diseases Research Program and the National Institutes of Health for funding this work. Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the true views of the Department of the Army or the Department of Defense.
Funding
B.W.C., M.G.T., A.C.J., E.A.A.-A., and D.V.Z. were supported by W0136_15_WR, a grant from the Military Infectious Diseases Research Program. W.M.H., R.J.M., and C.M. were supported by NIH Grant GM055769.
Footnotes
The authors declare no competing financial interest.
References
- 1.Boucher HW, Talbot GH, Benjamin DK, Jr, Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D. 10 × ’20 Progress—development of new drugs active against Gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2013;56:1685–1694. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kostyanev T, Bonten MJ, O’Brien S, Steel H, Ross S, François B, Tacconelli E, Winterhalter M, Stavenger RA, Karlén A, Harbarth S, Hackett J, Jafri HS, Vuong C, MacGowan A, Witschi A, Angyalosi G, Elborn JS, deWinter R, Goossens H. The Innovative Medicines Initiative’s New Drugs for Bad Bugs programme: European public-private partnerships for the development of new strategies to tackle antibiotic resistance. J. Antimicrob. Chemother. 2016;71:290. doi: 10.1093/jac/dkv339. [DOI] [PubMed] [Google Scholar]
- 3.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 4.Arduino SM, Quiroga MP, Ramirez MS, Merkier AK, Errecalde L, Di Martino A, Smayevsky J, Kaufman S, Centron D. Transposons and integrons in colistin-resistant clones of Klebsiella pneumoniae and Acinetobacter baumannii with epidemic or sporadic behaviour. J. Med. Microbiol. 2012;61:1417–1420. doi: 10.1099/jmm.0.038968-0. [DOI] [PubMed] [Google Scholar]
- 5.Dimopoulos G, Koulenti D, Tabah A, Poulakou G, Vesin A, Arvaniti K, Lathyris D, Matthaiou DK, Armaganidis A, Timsit JF. Bloodstream infections in ICU with increased resistance: epidemiology and outcomes. Minerva Anestesiol. 2015;81:405–418. [PubMed] [Google Scholar]
- 6.Behnia M, Logan SC, Fallen L, Catalano P. Nosocomial and ventilator-associated pneumonia in a community hospital intensive care unit: a retrospective review and analysis. BMC Res. Notes. 2014;7:232. doi: 10.1186/1756-0500-7-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnin RA, Poirel L, Nordmann P. New Delhi metallo-beta-lactamase-producing Acinetobacter baumannii: a novel paradigm for spreading antibiotic resistance genes. Future Microbiol. 2014;9:33–41. doi: 10.2217/fmb.13.69. [DOI] [PubMed] [Google Scholar]
- 8.Gupta SD, Gable K, Han G, Borovitskaya A, Selby L, Dunn TM, Harmon JM. Tsc10p and FVT1: topologically distinct short-chain reductases required for long-chain base synthesis in yeast and mammals. J. Lipid Res. 2009;50:1630–1640. doi: 10.1194/jlr.M800580-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snitkin ES, Zelazny AM, Montero CI, Stock F, Mijares L, Murray PR, Segre JA. Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13758–13763. doi: 10.1073/pnas.1104404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahl JW, Johnson JK, Harris AD, Phillippy AM, Hsiao WW, Thom KA, Rasko DA. Genomic comparison of multi-drug resistant invasive and colonizing Acinetobacter baumannii isolated from diverse human body sites reveals genomic plasticity. BMC Genomics. 2011;12:291. doi: 10.1186/1471-2164-12-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F, Zhu Y, Yi Y, Lu N, Zhu B, Hu Y. Comparative genomic analysis of Acinetobacter baumannii clinical isolates reveals extensive genomic variation and diverse antibiotic resistance determinants. BMC Genomics. 2014;15:1163. doi: 10.1186/1471-2164-15-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdulkareem AF, Lee HH, Ahmadi M, Martinez LR. Fungal serotype-specific differences in bacterial-yeast interactions. Virulence. 2015;6:654–659. doi: 10.1080/21505594.2015.1066962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhamgaye S, Murray G, Peleg AY. The influence of bacterial interaction on the virulence of Cryptococcus neoformans. Virulence. 2015;6:677–678. doi: 10.1080/21505594.2015.1088632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaddy JA, Tomaras AP, Actis LA. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 2009;77:3150–3160. doi: 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Touchon M, Cury J, Yoon EJ, Krizova L, Cerqueira GC, Murphy C, Feldgarden M, Wortman J, Clermont D, Lambert T, Grillot-Courvalin C, Nemec A, Courvalin P, Rocha EP. The genomic diversification of the whole Acinetobacter genus: origins, mechanisms, and consequences. Genome Biol. Evol. 2014;6:2866–2882. doi: 10.1093/gbe/evu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright MS, Stockwell TB, Beck E, Busam DA, Bajaksouzian S, Jacobs MR, Bonomo RA, Adams MD. SISPA-Seq for rapid whole genome surveys of bacterial isolates. Infect., Genet. Evol. 2015;32:191–198. doi: 10.1016/j.meegid.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, Huse HK, Zurawski DV, Brittnacher MJ, Manoil C. Resources for Genetic and Genomic Analysis of Emerging Pathogen Acinetobacter baumannii. J. Bacteriol. 2015;197:2027–2035. doi: 10.1128/JB.00131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin CE, Oh CS, Jiang Y. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids. 2007;1771:271–285. doi: 10.1016/j.bbalip.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 20.De Smet CH, Vittone E, Scherer M, Houweling M, Liebisch G, Brouwers JF, de Kroon AI. The yeast acyltransferase Sct1p regulates fatty acid desaturation by competing with the desaturase Ole1p. Mol. Biol. Cell. 2012;23:1146–1156. doi: 10.1091/mbc.E11-07-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu D, Sillaots S, Davison J, Hu W, Jiang B, Kauffman S, Martel N, Ocampo P, Oh C, Trosok S, Veillette K, Wang H, Yang M, Zhang L, Becker J, Martin CE, Roemer T. Chemical genetic profiling and characterization of small-molecule compounds that affect the biosynthesis of unsaturated fatty acids in Candida albicans. J. Biol. Chem. 2009;284:19754–19764. doi: 10.1074/jbc.M109.019877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbe V, Vallenet D, Fonknechten N, Kreimeyer A, Oztas S, Labarre L, Cruveiller S, Robert C, Duprat S, Wincker P, Ornston LN, Weissenbach J, Marlière P, Cohen GN, Médigue C. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 2004;32:5766–5779. doi: 10.1093/nar/gkh910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, Owen MT, Hallock JD, Kwak YI, Summers A, Li CZ, Rasko DA, Penwell WF, Honnold CL, Wise MC, Waterman PE, Lesho EP, Stewart RL, Actis LA, Palys TJ, Craft DW, Zurawski DV. AB5075, a Highly Virulent Isolate of Acinetobacter baumannii, as a Model Strain for the Evaluation of Pathogenesis and Antimicrobial Treatments. mBio. 2014;5:e01076–14. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang R, Wang Y, Tan L, Zhang HY, Yang M. Analysis of polysorbate 80 and its related compounds by RP-HPLC with ELSD and MS detection. J. Chromatogr. Sci. 2012;50:598–607. doi: 10.1093/chromsci/bms035. [DOI] [PubMed] [Google Scholar]
- 25.Weiss A, Broach WH, Lee MC, Shaw LN. Towards the complete small RNome of Acinetobacter baumannii. mGen. 2016;2:3. doi: 10.1099/mgen.0.000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C, Guo ZB, Du ZM, Yang HY, Bi YJ, Wang GQ, Tan YF. Cellular fatty acids as chemical markers for differentiation of Acinetobacter baumannii and Acinetobacter calcoaceticus. Biomed. Environ. Sci. 2012;25:711–717. doi: 10.3967/0895-3988.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 27.Lu H, Tonge PJ. Inhibitors of FabI, an enzyme drug target in the bacterial fatty acid biosynthesis pathway. Acc. Chem. Res. 2008;41:11–20. doi: 10.1021/ar700156e. [DOI] [PubMed] [Google Scholar]
- 28.Parsons JB, Frank MW, Subramanian C, Saenkham P, Rock CO. Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15378–15383. doi: 10.1073/pnas.1109208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsons JB, Rock CO. Bacterial lipids: metabolism and membrane homeostasis. Prog. Lipid Res. 2013;52:249–276. doi: 10.1016/j.plipres.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan Y, Leeds JA, Meredith TC. Pseudomonas aeruginosa directly shunts beta-oxidation degradation intermediates into de novo fatty acid biosynthesis. J. Bacteriol. 2012;194:5185–5196. doi: 10.1128/JB.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]