Figure 4.
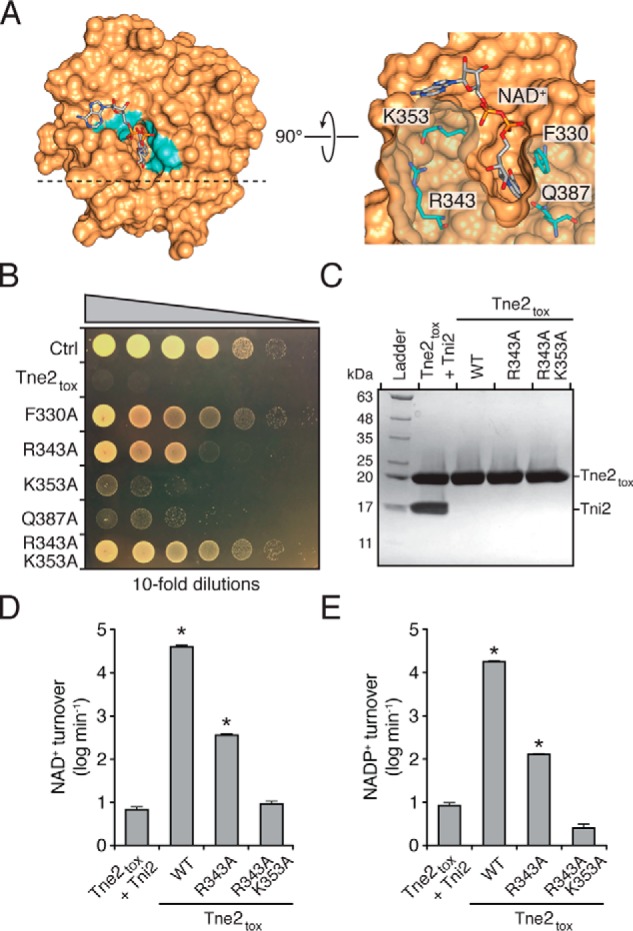
Identification of residues required for optimal NAD(P)+ hydrolysis by Tne2tox. A, molecular docking places NAD+ in the predicted active-site pocket of Tne2tox. Phe-330, Arg-343, Lys-353, and Gln-413, which are conserved among Tne2 orthologous proteins and line the NAD+-binding pocket, are indicated. B, growth of E. coli cells expressing a vector control (Ctrl), Tne2tox, or the indicated site-specific variants of Tne2tox. C, SDS-PAGE analysis of the Tne2tox–Tni2 complex and refolded Tne2tox and the indicated site-specific mutants thereof. Tne2tox and its variants possess an N-terminal hexahistidine tag, whereas Tni2 is untagged. Proteins were visualized using Coomassie Blue staining. D and E, hydrolysis rates of NAD+ (D) and NADP+ (E) by the purified proteins indicated in C. Error bars represent ± S.D. (n = 3). Asterisks indicate NAD(P)+ hydrolysis rates that are significantly higher than the Tni2-inhibited control.
