Significance
Amplification of the cyclin E genes and overexpression of cyclin E proteins is very frequent in several human tumor types. In particular, overexpression of E cyclins was implicated in the pathogenesis of liver cancers. According to the current cell division models, E cyclins drive tumor cell proliferation by activating its kinase partner, the cyclin-dependent kinase 2 (CDK2). In this study, we demonstrate that E cyclins are dispensable for adult mouse physiology, but essential for liver cancer progression. Surprisingly, we found that the function of E cyclins in liver tumorigenesis is kinase independent. Our study suggests that agents targeting E cyclins would be efficacious in liver cancer treatment, as they would halt proliferation of tumor cells while sparing normal tissues.
Keywords: cell cycle, E-type cyclins, cyclin-dependent kinase CDK2, liver cancer
Abstract
E-type cyclins (cyclins E1 and E2) are components of the core cell cycle machinery and are overexpressed in many human tumor types. E cyclins are thought to drive tumor cell proliferation by activating the cyclin-dependent kinase 2 (CDK2). The cyclin E1 gene represents the site of recurrent integration of the hepatitis B virus in the pathogenesis of hepatocellular carcinoma, and this event is associated with strong up-regulation of cyclin E1 expression. Regardless of the underlying mechanism of tumorigenesis, the majority of liver cancers overexpress E-type cyclins. Here we used conditional cyclin E knockout mice and a liver cancer model to test the requirement for the function of E cyclins in liver tumorigenesis. We show that a ubiquitous, global shutdown of E cyclins did not visibly affect postnatal development or physiology of adult mice. However, an acute ablation of E cyclins halted liver cancer progression. We demonstrated that also human liver cancer cells critically depend on E cyclins for proliferation. In contrast, we found that the function of the cyclin E catalytic partner, CDK2, is dispensable in liver cancer cells. We observed that E cyclins drive proliferation of tumor cells in a CDK2- and kinase-independent mechanism. Our study suggests that compounds which degrade or inhibit cyclin E might represent a highly selective therapeutic strategy for patients with liver cancer, as these compounds would selectively cripple proliferation of tumor cells, while sparing normal tissues.
E-type cyclins (cyclins E1 and E2, collectively called “cyclin E”) are components of the core cell cycle machinery. These two cyclins are encoded by separate genes and are usually coexpressed in proliferating cells (1). E-type cyclins bind and activate their kinase partner, the cyclin-dependent kinase 2 (CDK2). Cyclin E–CDK2 complexes phosphorylate proteins governing cell cycle progression, including the retinoblastoma protein and retinoblastoma-related p107 and p130 proteins, as well as proteins involved in histone biosynthesis, centrosome duplication, and firing of DNA replication origins (2). These functions of cyclin E–CDK2 kinase promote progression of cells through the G1 phase of the cell cycle and entry into DNA synthesis (S phase) (2).
Consistent with their growth-promoting roles, amplification of the cyclin E1 and E2 genes (CCNE1 and CCNE2, respectively) and pathological overexpression of their proteins have been documented in many cancer types (3–7). Indeed, the CCNE1 gene located on chromosome 19q12 represents one of the most frequently amplified loci across all human tumor types (8). Overexpression of E cyclins was shown to generally correlate with poor clinical outcome (3, 4). In the case of HER2-positive breast cancers, cyclin E amplification/overexpression represents a molecular mechanism of resistance to trastuzumab (Herceptin) treatment (9). It is currently assumed that overexpressed E cyclins drive the tumorigenic process through hyperactivation of the CDK2 kinase.
Hepatocellular carcinoma (HCC) represents the second most common cause of death from cancer worldwide, and it is responsible for 745,000 deaths annually (10). The 5-y survival rate for patients with HCC is only ∼7%. E cyclins are overexpressed in the majority of HCC cases, while nearly 20% of tumors display amplification of the CCNE1 and CCNE2 genes (11–14). The major risk factors for HCC are chronic hepatitis B virus (HBV) or hepatitis C virus (HCV) infections (15). The CCNE1 gene represents one of the most frequent sites of HBV integration in HCC, and this event is associated with strong up-regulation of cyclin E1 expression (16). Moreover, the hepatitis C virus core protein was shown to promote cell proliferation by up-regulating cyclin E levels (17). Regardless of the pathogenesis, HCC occurs more than five times more frequently in males than in females, and this was attributed in part to up-regulation of cyclin E expression by testosterone (18). Collectively, these findings suggest an important role for cyclin E in pathogenesis of HCC.
In this study, we tested the requirement for E cyclins in progression of HCC using mouse cancer models. We report that E cyclins play an essential, rate-limiting role in liver cancer progression, while being dispensable for proliferation of normal tissues. Unexpectedly, we found that the function of E cyclins in liver cancer cell proliferation is independent of the canonical role of E cyclins as activators of CDK2.
Results
Cyclin E Function Is Dispensable in Postnatal Animals.
We and others previously reported that constitutive, germline ablation of cyclin E1 and E2 resulted in an embryonic lethality of cyclin E1−/−E2−/− mice due to placental and heart abnormalities (19, 20). To circumvent this lethality, and to study the requirement of cyclin E function at later stages of development, we generated a conditional cyclin E1 knockout (E1F/F) mouse strain (21). We interbred cyclin E1F/F and cyclin E2−/− mice giving rise to cyclin E1F/FE2−/− animals, which will be further referred to as “conditional cyclin E knockout” mice.
In the first set of experiments, we crossed conditional cyclin E knockout mice with Esr1-Cre animals (22), which ubiquitously express tamoxifen-inducible Cre recombinase. Administration of tamoxifen to Esr1-Cre mice activates Cre, leading to a global deletion of the “floxed” sequences (22). We administered tamoxifen to pregnant females bearing cyclin E1F/FE2−/− embryos at day 17.5 of gestation, and continued treatment of postnatal animals with tamoxifen to ensure ubiquitous deletion of E cyclins (Fig. S1A). We then verified an efficient deletion of cyclin E1 in most organs analyzed (Fig. S1B, see P36) and observed the animals for 1 y. We found that an acute and global shutdown of E cyclins did not compromise the animals’ viability and had no outward effect on their health, except for slightly reduced body weights of cyclin E-deleted animals (Fig. S1C). Cyclin E-deleted animals displayed normal biochemical parameters in the peripheral blood and normal parameters of liver function (Fig. S1 D and E). Importantly, efficient deletion of cyclin E1 was maintained after 1 y even in highly proliferative organs such as intestine or spleens (Fig. S1B, see 1 y). We concluded that E cyclins are largely dispensable for proliferation of cells in postnatal animals. Despite the efficient deletion of cyclin E1 after Cre activation, we detected low levels of nondeleted floxed cyclin E alleles in many organs (Fig. S1B). Hence, we cannot conclude that all cells within a given compartment do not require cyclin E.
Requirement for Cyclin E Function in Liver Cancer Progression.
We next asked what the impact would be of cyclin E shutdown on liver tumorigenesis. To efficiently delete both E cyclins in livers, we turned to Mx1-Cre mice, which express inducible Cre recombinase. Administration of a double-stranded RNA analog polyI–polyC to Mx1-Cre animals activates Cre, leading to nearly 100% deletion of the floxed sequences in liver and in the bone marrow cells (Fig. S2 A and B) (23). First, we administered polyI–polyC to cyclin E1F/FE2−/−/Mx1-Cre animals and determined that ablation of both E cyclins had no effect on hematological parameters (Fig. S2 C and D). These findings are consistent with the observation that bone marrow cells express very low levels of E cyclins (24).
To induce liver tumorigenesis, we injected 2-wk-old male mice with a chemical carcinogen diethylnitrosamine (DEN) (Fig. 1A). It is well established that this induces HCC formation in essentially all male mice, and that tumors recapitulate gender disparity, histopathological appearance, and genetic signature of human HCC (25). We verified that DEN-induced mouse HCC tumors expressed high levels of E cyclins (Fig. 1B).
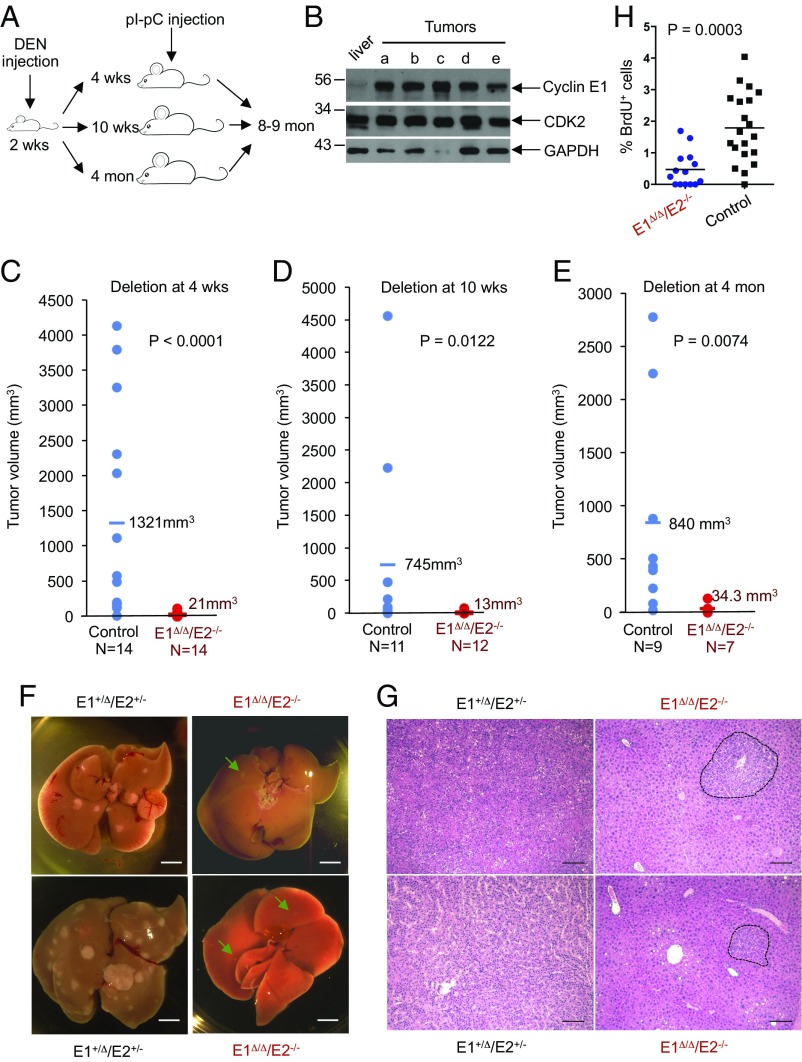
Fig. 1.
E-type cyclins are required for liver cancer progression. (A) Experimental outline. Diethylnitrosamine (DEN) was used to trigger development of hepatocellular carcinoma; injection of polyinosinic–polycytidylic acid (pI–pC) was used to induce Cre recombinase and delete E-type cyclins. Male mice of the following genotypes were used: cyclin E1+/FE2+/−/Mx1-Cre (control) and cyclin E1F/FE2−/−/Mx1-Cre (experimental). (B) Western blot analysis of cyclin E1 levels in five liver tumors (a–e) collected from DEN-injected 8-mo-old mice. For comparison, normal liver was also analyzed. Note that cyclin E level is very low in normal liver but quite high in liver tumors. (C) The estimated tumor burden (per mouse) in control cyclin E1+/ΔE2+/−/Mx1-Cre and in cyclin E1Δ/ΔE2−/−/Mx1-Cre (cyclin E-deleted) mice, 8–9 mo after DEN injection. Mice were injected with pI–pC (to delete cyclin E) at 4 wk of age. Each dot corresponds to an individual mouse; horizontal lines depict mean values. (D) Similar analysis as in C, except that cyclin E was deleted at 10 wk. (E) Similar analysis as in C, cyclin E deletion at 4 mo. In C–E, P values were calculated using Kolmogorov–Smirnov test. (F) Representative images of livers with tumors in control cyclin E1+/ΔE2+/−/Mx1-Cre and in cyclin E-deleted (cyclin E1Δ/ΔE2−/−/Mx1-Cre) mice, 8 mo after DEN injection. Cyclin E was deleted (through pI–pC administration) at 4 mo. Green arrows point to small tumors in cyclin E-deleted mice. (Scale bar, 10 mm.) (G) Sections of livers with tumors, as in F, stained with hematoxylin and eosin. In sections from cyclin E1+/ΔE2+/−/Mx1-Cre livers, tumors occupy the entire field of view; dashed lines indicate boundaries of small tumors in cyclin E1Δ/ΔE2−/−/Mx1-Cre mice. (Scale bar, 100 μm.) (H) Quantification of BrdU-positive cells in sections of liver tumors from cyclin E1+/ΔE2+/−/Mx1-Cre (control, 20 tumors from four mice) and cyclin E-deleted (E1Δ/ΔE2−/−/Mx1-Cre mice; 14 tumors from four mice). Mice were injected with pI–pC three times at 4 mo after DEN injection and killed 7 d after the last pI–pC dose. P value was calculated using two-tailed t test.
In the first experiments, we ablated both E cyclins (through administration of polyI–polyC) at 4 wk of age, i.e., 2 wk after DEN administration, and killed the animals after 8 mo (Fig. 1A). As expected, control polyI–polyC-treated cyclin E1+/FE2+/−/Mx1-Cre mice displayed massive liver tumors. In striking contrast, cyclin E-deleted mice developed only a few very small tumors (Fig. 1C).
In the next set of experiments, we injected male neonates with DEN, as above, and allowed animals to develop microscopic tumors for 8 wk (Fig. 1A). Tumor-bearing mice were then injected with polyI–polyC, thereby triggering ablation of both E cyclins. Mice were observed until they reached 8–9 mo of age, when livers were collected and analyzed. Again, control mice developed large liver tumors. In contrast, in cyclin E-deleted mice, tumor progression was halted and they developed only a few very small tumors (Fig. 1D).
Lastly, we ablated E cyclins at 4 mo post-DEN administration, i.e., after the animals had developed macroscopic tumors, and collected the livers at 8–9 mo of age (Fig. 1A). We found that ablation of both E cyclins arrested tumor progression at the stage of very small tumors (Fig. 1 E–G). We also compared tumor sizes at 4 mo, soon after polyI–polyC administration, versus at the end of observation period, i.e., at 8–9 mo of age. As expected, control tumors grew considerably during this period. In contrast, the growth of tumors was completely halted upon cyclin E ablation (Fig. S3A). In addition, we injected mice with BrdU and quantified tumor cell proliferation (BrdU incorporation) as well as the apoptotic rate (TUNEL staining). We found that ablation of both E cyclins strongly decreased proliferation of tumor cells (Fig. 1H). On the other hand, we observed no apoptosis in control or cyclin E-deleted tumors (Fig. S3B).
Collectively, these findings indicate that E cyclins play an essential and rate-limiting role in HCC progression, by driving proliferation of liver cancer cells. In contrast, cyclin E function is not required for proliferation of cells in several normal nontransformed tissues.
E Cyclins Are Required for Proliferation of Human HCC Cells.
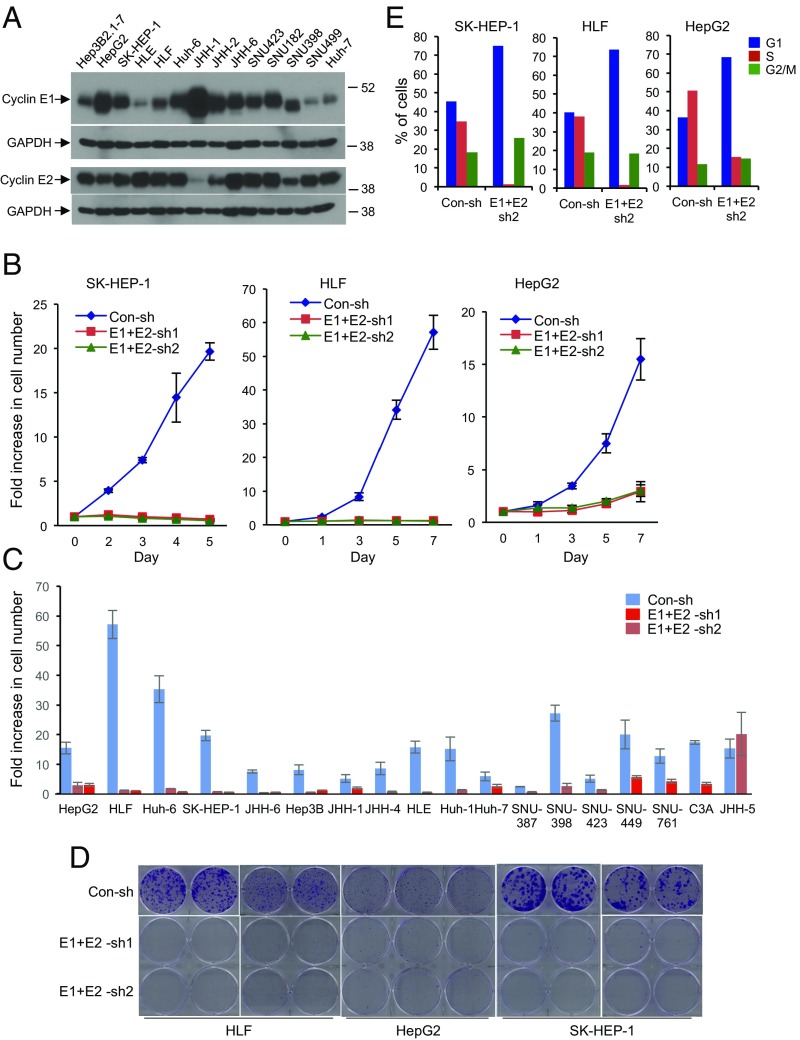
We next asked whether this requirement for the function of E cyclins also operates in human HCC cells. To address this question, we collected 20 human HCC cell lines and established that all of them express high levels of cyclin E1 and/or E2 (Fig. 2A and Fig. S4A). We then depleted both cyclins E1 and E2, using several independent shRNAs, in all 20 human HCC cell lines (Fig. S4B). We found that depletion of both E-type cyclins essentially extinguished proliferation of 19 of the 20 HCC cell lines (Fig. 2 B and C). Knockdown of E cyclins also abrogated the ability of HCC cells to form colonies (Fig. 2D). Flow cytometric analyses revealed that depletion of cyclins E1 and E2 led to a significant decline in the fraction of S phase cells, consistent with a likely proliferative arrest of tumor cells (Fig. 2E and Fig. S4C). On the other hand, we detected no increase in the apoptotic rate of HCC cells after cyclin E depletion (Fig. S5A). We concluded that E cyclins are essential for proliferation also in human liver cancer cells.
Fig. 2.
E-type cyclins are required for human hepatocellular carcinoma (HCC) cell proliferation. (A) Western blot analysis of cyclin E1 and E2 levels in the indicated human HCC cell lines. GAPDH was used as a loading control. (B) Growth curves of human HCC cell lines SK-HEP-1, HLF, and HepG2. Cells were transduced with viruses encoding two different sets of anti-cyclin E1 and E2 shRNAs (E1+E2-sh1, E1+E2-sh2), or control shRNA (Con-sh). Error bars indicate SD, n = 3. (C) Fold increase in cell numbers in the indicated 18 human HCC cell lines following depletion of cyclins E1 and E2. Cells were transduced as in B, plated, and the fold increase in cell number was measured 6–7 d after plating. Note that only in JHH-5 cells, the growth rate was not affected by depletion of cyclins E1 and E2. Error bars indicate SD, n = 3. (D) Clonogenicity assays of HLF, HepG2, and SK-HEP-1 cells following depletion of cyclins E1 and E2, as in B. (E) Cell cycle distribution of human HCC cell lines SK-HEP-1, HLF, and HepG2 following depletion of cyclins E1 and E2. Cells were pulsed with bromodeoxyuridine (BrdU), stained with an anti-BrdU antibody and propidium iodide, and analyzed by flow cytometry. The percentages of cells in each cell cycle phase are indicated.
E Cyclins Drive HCC Proliferation via a Kinase-Independent Mechanism.
E cyclins are thought to drive tumor cell proliferation through activation of their kinase partner, CDK2. We verified that in human HCC cells, cyclin E forms a stoichiometric complex with CDK2. Specifically, immunodepletion of CDK2 removed essentially all cyclin E from HCC cell lysates (Fig. 3A). In contrast, we did not detect interaction of cyclin E with a related kinase, CDK1, and immunodepletion of CDK1 from HCC cells had no effect on cyclin E levels (Fig. 3A).
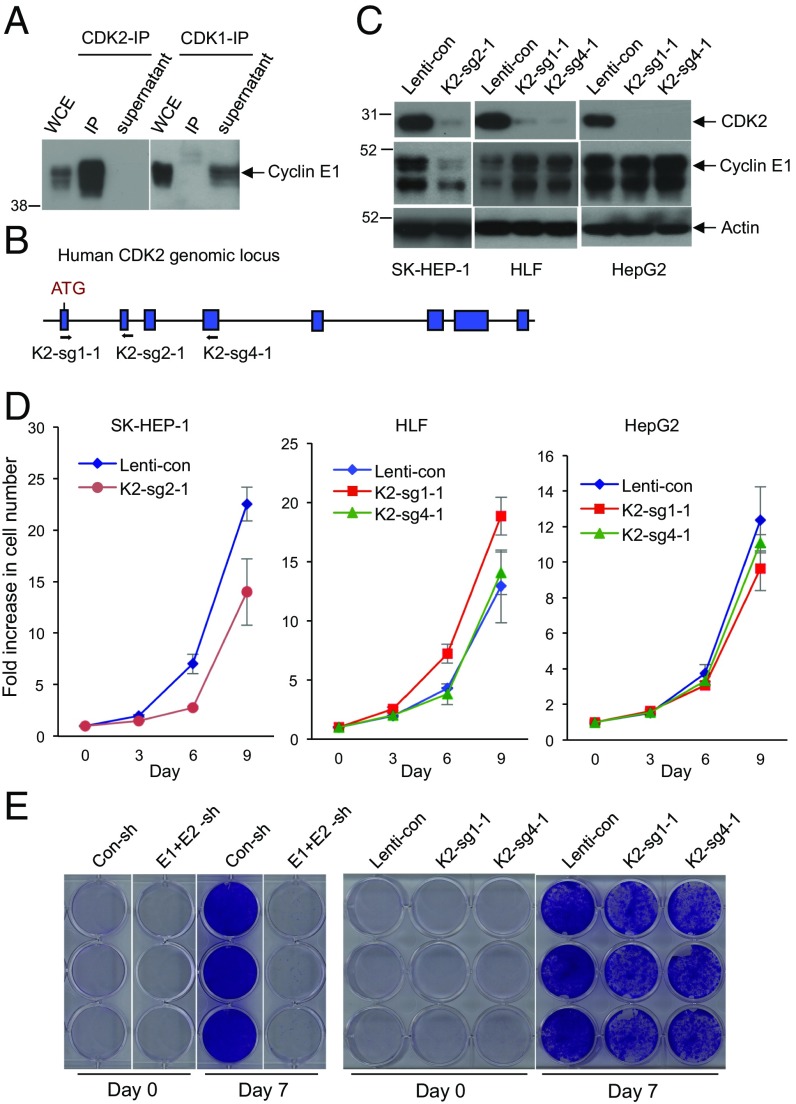
Fig. 3.
CDK2 is dispensable for HCC cell proliferation. (A) Immunoprecipitation (IP) using anti-CDK2 (Left) or anti-CDK1 (Right) antibodies, followed by Western blotting with an anti-cyclin E1 antibody. Note that immunoprecipitated CDK2 brings down large amounts of cyclin E1, and that there is essentially no cyclin E1 left in the supernatant after CDK2-IP. In contrast, no cyclin E1 was detected in CDK1-IP, and cyclin E1 remained in the supernatant after CDK1-IP. Whole cell extract (WCE) was also immunoblotted. (B) Design scheme of three independent guide RNAs against human CDK2. (C) Western blot analysis of CDK2 levels in three HCC cell lines after CRISPR-mediated knockout of CDK2 using three independent guide RNAs (K2-sg2-1, K2-sg1-1, and K2-sg4-1). Actin was used as a loading control. (D) Growth curves of SK-HEP-1, HLF, and HepG2 liver cancer cells following transduction with lentiviruses encoding K2-sg2-1, K2-sg1-1, or K2-sg4-1 guide RNAs (CDK2-knockout cells) compared with cells transduced with control viruses (Lenti-con). Error bars indicate SD, n = 3. (E) Comparison of HCC Huh-6 cell growth following knockdown of cyclins E1 and E2 (E1+E2-sh) versus after CRISPR-mediated knockout of CDK2 (K2-sg1-1 and K2-sg4-1). Con-sh and Lenti-con denote cells transduced with control shRNA or control-Lenti-viruses, respectively. Cells were fixed and stained with crystal violet 7 d after plating.
To test the requirement for CDK2 function in HCC proliferation, we used CRISPR/Cas9 to knock out CDK2 in four human HCC cell lines (Fig. 3 B and C). Unexpectedly, we found that CDK2-null HCC cells proliferated nearly normally (Fig. 3 D and E). This was in contrast to depletion of E cyclins, which arrested cell growth (Figs. 2 B–E and 3E). These observations suggested that E cyclins may play a CDK2-independent role in driving HCC proliferation.
It was possible that upon ablation of CDK2, another CDK forms a complex with E cyclins, thereby replacing CDK2 function. Indeed, E cyclins can also interact with CDK1 and CDK3 in certain conditions (26). To explore this possibility, we immunoprecipitated cyclin E from CDK2-knockout HCC cells and gauged cyclin E-associated kinase activity by performing in vitro kinase assays. We detected essentially no cyclin E-associated kinase activity in CDK2-null cells (Fig. 4A), arguing against compensatory replacement of CDK2 function by a related kinase.
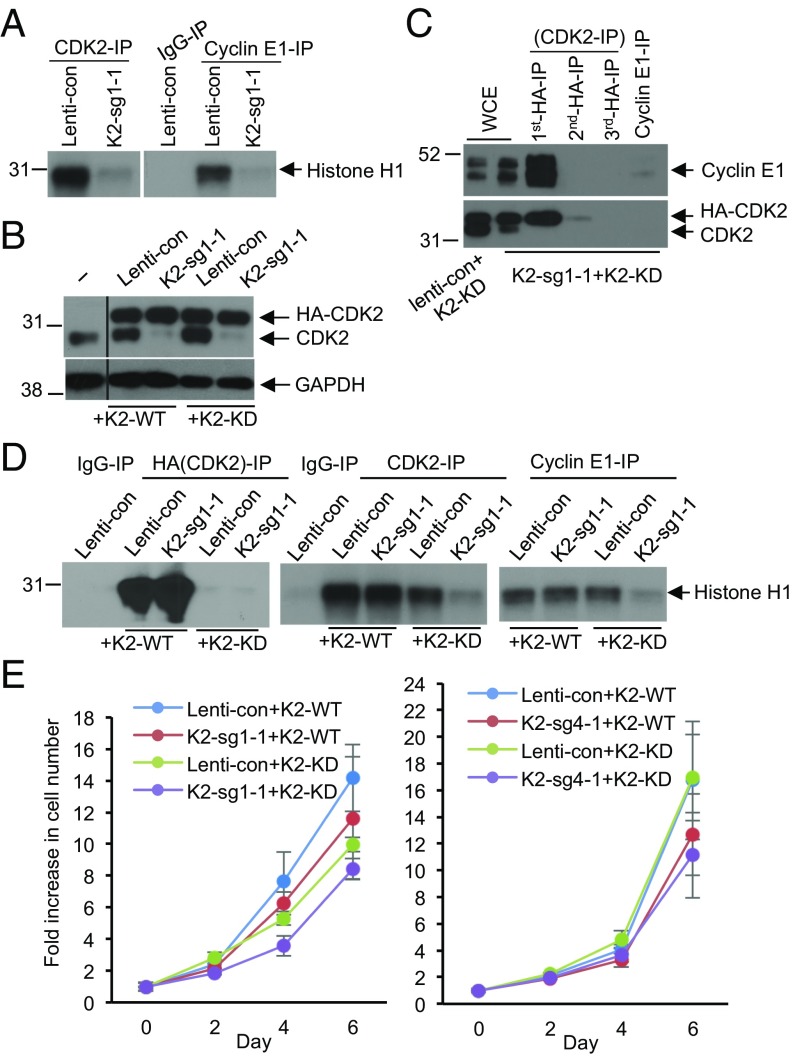
Fig. 4.
Cyclin E–CDK2 kinase activity is not required for proliferation of HCC cells. (A) CDK2-associated kinase activity (Left) and cyclin E1-associated kinase activity (Right) after CRISPR-mediated knockout of CDK2 in HepG2 cells. CDK2 or cyclin E1 were immunoprecipitated (IP) from control cells (Lenti-con) or from CDK2-knockout cells (K2-sg1-1), and used for in vitro kinase reactions with histone H1 as substrate. For control, immunoprecipitation with IgG (IgG-IP) was used. (B) Western blot analysis of the expression levels of the endogenous CDK2 and ectopically expressed HA-tagged wild-type or kinase-dead CDK2 in parental HepG2 cells (−), or cells transduced with control virus (Lenti-con), or in CDK2-knockout cells (K2-sg1-1) ectopically expressing wild type CDK2 (+K2-WT) or kinase-dead CDK2 (+K2-KD). GAPDH was used as loading control. The middle portion of the blot was spliced out (indicated by a line). (C) IP-Western blot analysis of the interaction between the endogenous cyclin E1 and kinase-dead CDK2 in CDK2-knockout HepG2 cells (K2-sg1-1) ectopically expressing HA-tagged kinase-dead CDK2 (+K2-KD). The successive rounds of IP were performed with an anti-HA antibody, the supernatant after the second round was immunoprecipitated using an anti-cyclin E1 antibody, and the immunoblots were probed with an anti-cyclin E1 antibody (Upper) and CDK2 antibody (Lower). WCE, whole cell extracts. (D) Kinase assays to measure the catalytic activity of the ectopically expressed wild-type and kinase-dead CDK2 (anti-HA IP, Left), or total CDK2 kinase activity (anti-CDK2 IP, Middle), or cyclin E1-associated kinase activity (anti-cyclin E1-IP, Right) in control cells (Lenti-con) or CDK2-knockout cells (K2-sg1-1) ectopically expressing either wild-type CDK2 (+K2-WT) or kinase-dead CDK2 (+K2-KD). Histone H1 was used as substrate. IgG-IP served as a negative control. (E) Growth curves of control (Lenti-con) or CDK2-knockout cells (K2-sg1-1 and K2-sg4-1) ectopically expressing either wild-type CDK2 (+K2-WT) or kinase-dead CDK2 (+K2-KD). The two panels represent two HCC CDK2-knockout cell lines obtained using two independent guide RNAs against human CDK2 (Left, K2-sg1-1 in HepG2 cells and Right, K2-sg4-1 in HLF cells). Error bars indicate SD, n = 3.
To further address this possibility, we ectopically expressed kinase-dead CDK2 mutant (CDK2KD) in CDK2-null cells (Fig. 4B). We established that in these cells, essentially all cyclin E was bound to CDK2KD, as immunodepletion of CDK2KD fully depleted cyclin E protein (Fig. 4C). As expected, immunoprecipitation of cyclin E followed by in vitro kinase assays revealed virtually no cyclin E-associated kinase in cells expressing kinase-dead CDK2 (Fig. 4D). Strikingly, HCC cells expressing kinase-dead cyclin E–CDK2KD complexes proliferated nearly normally, indicating that catalytically inactive cyclin E can drive proliferation of HCC cells (Fig. 4E). These observations further reinforced our conclusion that E cyclins drive HCC proliferation via a kinase-independent mechanism.
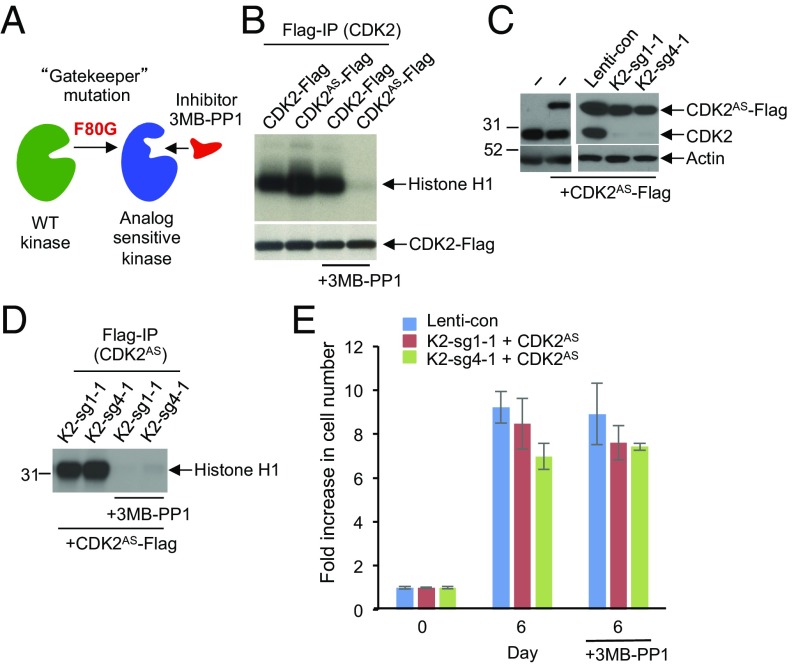
We extended these analyses using analog-sensitive CDK2. In the analog-sensitive approach, the large hydrophobic residue in the kinase active site, called the “gatekeeper,” is mutated from the naturally occurring bulky residue to glycine or alanine. This creates an enlarged pocket not found in any wild-type kinase (27–29). The engineered kinase, termed “analog sensitive,” can be potently and uniquely inhibited by inhibitors that occupy this enlarged ATP-binding pocket, such as 3MB-PP1 (Fig. 5A). Importantly, 3MB-PP1 does not inhibit any wild-type kinases in the mammalian kinome (27–29). We engineered analog-sensitive CDK2 (CDK2AS) and verified that it can be potently inhibited by 3MB-PP1 (Fig. 5B).
Fig. 5.
Analyses of HCC cells expressing analog-sensitive CDK2. (A) Diagram illustrating the principle of analog-sensitive kinases. (B) Flag-tagged wild-type or analog-sensitive CDK2 (CDK2AS) were ectopically expressed in 293T cells, immunoprecipitated, and subjected to in vitro kinase assays in the presence or absence of 1 μM 3MB-PP1. Note inhibition of the analog-sensitive CDK2 by 3MB-PP1. (C) Western blot analysis of the levels of ectopically expressed analog-sensitive CDK2 (CDK2AS-Flag) and the endogenous CDK2 (CDK2) in parental HepG2 cells (−), or HepG2 cells transduced with control lenti-viruses (Lenti-con), or in CDK2-knockout HepG2 cells (K2-sg1-1 and K2-sg4-1). Actin was used as loading control. (D) CDK2-knockout HepG2 cells (K2-sg1-1 and K2-sg4-1) were engineered to ectopically express Flag-tagged analog-sensitive CDK2 (+CDK2AS-Flag). CDK2AS was immunoprecipitated with an anti-Flag antibody and used for in vitro kinase assays using histone H1 as a substrate. Note that treatment with 3MB-PP1 inhibited the catalytic activity of CDK2AS. (E) Cell growth of control HepG2 cells (Lenti-con) or CDK2-knockout HepG2 cells (K2-sg1-1 and K2-sg4-1) ectopically expressing analog-sensitive CDK2 (+CDK2AS). Cells were cultured in the presence or absence of 3MB-PP1, and fold increase in cell numbers was determined after 6 d. Shown are mean values, error bars indicate SD, n = 3.
We next ectopically expressed CDK2AS in CDK2-null HCC cells (Fig. 5C). As expected, CDK2AS was catalytically active in the absence of 3MB-PP1. In contrast, treatment of cells with 3MB-PP1 inhibited CDK2AS kinase activity (Fig. 5D). Importantly, cells expressing cyclin E–CDK2AS complexes proliferated normally in the presence of 3MB-PP1 (Fig. 5E), indicating that kinase-inactive cyclin E–CDK2AS complexes can drive tumor cell proliferation.
Lastly, we took advantage of the cyclin E1188-192A mutant that retains CDK2 binding, but it is deficient in activating CDK2 kinase (30). We depleted E cyclins in HCC cells, and reexpressed shRNA-resistant cyclin E1188-192A. This kinase-deficient mutant largely rescued the proliferative deficiency of cyclin E-depleted cells (Fig. S5 B and C). Collectively, these observations indicate that E-type cyclins drive proliferation of human liver cancer cells via a kinase-independent mechanism.
Discussion
Cyclin E represents a component of the core cell cycle machinery. We and others previously showed that combined ablation of cyclins E1 and E2 resulted in an embryonic lethality (19, 20). In this study, we bypassed the requirement for cyclin E function during embryogenesis by ubiquitously ablating both E cyclins in utero at the very end of gestation. We report here that E cyclins are largely dispensable for postnatal development and for normal physiology of adult animals. In contrast, we found that E cyclins are critically required for progression of liver cancers.
It is not clear why only tumor cells depend on cyclin E function. However, similar dependence on individual cell cycle proteins has been documented for other tumor types. For instance, inducible ablation of CDK4 specifically affected proliferation of RasG12V-driven non-small cell lung cancers (31). Likewise, an acute and global ablation of cyclin D1, or chemical inhibition of CDK4 and CDK6 kinase in mice bearing HER2-driven mammary carcinomas, halted proliferation of breast cancer cells without having any obvious effects on normal nontransformed tissues (32). It seems that particular tumor types critically require individual cyclin proteins, depending on genetic lesions they carry, while in normal cells the cell cycle machinery operates in a more “plastic” and redundant mode. This specific requirement for individual cyclins and CDKs is now being translated into novel anticancer therapies. Thus, inhibitors of CDK4 and CDK6 palbociclib, ribociclib, and abemaciclib received “breakthrough therapy” designation status from the Food and Drug Administration and are currently being used in clinical trials for patients with breast cancer with very promising results (33–35).
According to the current cell division models, E cyclins contribute to tumor cell proliferation by activating the kinase activity of their catalytic partner, CDK2. As described above, we observed that cells expressing stoichiometric “kinase-dead” cyclin E–CDK2 complexes proliferated normally, which we interpreted as an indication that cyclin E-associated kinase activity is largely dispensable for proliferation of HCC cells. Indeed, E cyclins were previously postulated to perform CDK2- and kinase-independent functions, by physically localizing to the centrosomes (36), or by facilitating loading of MCM replicative helicase in cells exiting quiescence (37). It is unclear, however, whether these observations can explain the rate-limiting, kinase-independent role of cyclin E specifically in tumor cells. Further studies are needed to conclusively address this point.
It should be noted that, at least during mouse embryonic development, all cyclins (including cyclin E) can bind Cdk1 (38). Moreover, Cdk1 is sufficient to drive embryonic development in the absence of other interphase Cdks (38). Hence, it is possible that also in human cancer cells, cyclin E associates with CDK1 and plays a role in driving tumor cell proliferation.
Regardless of the molecular role of E cyclins in HCC, our work presented here suggests that compounds that target E cyclins, but not inhibitors of cyclin E–CDK2 kinase, might be highly efficacious in treatment of liver cancers. Recent development of phthalimide-based bivalent compounds (39) makes it possible to design compounds that selectively degrade E cyclins. We predict that such agents might represent a selective therapeutic strategy for patients with liver cancer, as these compounds might cripple proliferation of tumor cells, while sparing normal tissues.
Materials and Methods
Experimental Animals.
All mouse experiments were approved by the Dana-Farber Cancer Institute Animal Care and Use Committee.
Conditional cyclin E1 knockout mice were generated as described (21). Cyclin E1F/FE2−/− mice were further bred with Esr1-Cre and Mx1-Cre mice (from The Jackson Laboratory).
Cell Culture.
Human hepatocellular carcinoma cell lines were obtained either from the Japan Cell Bank or from ATCC. All cells were cultured according to the recommendation of the suppliers. All JHH lines were established by Seishi Nagamori at National Institute of Infectious Diseases, Japan.
Viral Transduction.
The following lentiviral vectors were used for viral transduction: pLKO-puro vector containing shRNAs against cyclin E1 (A8: 5′-GCAATTCTTCTGGATTGGTTA-3′; A9: 5′-CGACATAGAGAACTGTGTCAA-3′), and cyclin E2 (A1: 5′-GCTCTTAAAGATGCTCCTAAA-3′; A2: 5′-CCAGACACATACAAACTATTT-3′) (from RNAi Consortium https://www.broadinstitute.org/rnai/trc).
Single-strand guide RNAs against CDK2 were designed using a software Target Finder from Feng Zhang’s laboratory at Broad Institute: Sg1-1 5′-CAGAAACAAGTTGACGGGAG-3′, Sg4-1 5′-TCTGAGGTTTAAGGTCTCGG-3′, and Sg2-1 5′-GATCTCTCGGATGGCAGTAC-3′ and cloned into pLentiCrispr-v2-puro vector (Addgene).
Western Blotting.
Whole cell extracts were obtained and proteins were solubilized for immunoblotting as described (40). Antibodies against the following proteins were used: cyclin E1 (sc-481, 1:500; Santa Cruz and sc-247, 1:500; Santa Cruz), CDK2 (sc-163, 1:1,000; Santa Cruz), cyclin E2 (EP454Y, 1:1,000; Millipore), CDK1 (ab71939, 1:1,000; Abcam), GAPDH (D16H11, 1:1,000; Cell Signaling), and actin (AC40, 1:3,000; Sigma).
Supplementary Material
Acknowledgments
We thank Drs. Junko Odajima, Tobias Otto from the P.S. laboratory, and Ms. Li Zhang from Rodent Histopathology Core for help. This study was supported by NIH R01 CA202634, R01 CA083688, and R01 CA132740 (to P.S.).
Footnotes
Conflict of interest statement: P.S. is a consultant and a recipient of a research grant from Novartis.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711477115/-/DCSupplemental.
References
- 1.Geng Y, et al. Expression of cyclins E1 and E2 during mouse development and in neoplasia. Proc Natl Acad Sci USA. 2001;98:13138–13143. doi: 10.1073/pnas.231487798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–2786. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 3.Gao S, Ma JJ, Lu C. Prognostic value of cyclin E expression in breast cancer: A meta-analysis. Tumour Biol. 2013;34:3423–3430. doi: 10.1007/s13277-013-0915-8. [DOI] [PubMed] [Google Scholar]
- 4.Nakayama K, et al. CCNE1 amplification is associated with aggressive potential in endometrioid endometrial carcinomas. Int J Oncol. 2016;48:506–516. doi: 10.3892/ijo.2015.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patch AM, et al. Australian Ovarian Cancer Study Group Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 6.Schraml P, et al. Cyclin E overexpression and amplification in human tumours. J Pathol. 2003;200:375–382. doi: 10.1002/path.1356. [DOI] [PubMed] [Google Scholar]
- 7.Shaye A, et al. Cyclin E deregulation is an early event in the development of breast cancer. Breast Cancer Res Treat. 2009;115:651–659. doi: 10.1007/s10549-008-0266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scaltriti M, et al. Cyclin E amplification/overexpression is a mechanism of trastuzumab resistance in HER2+ breast cancer patients. Proc Natl Acad Sci USA. 2011;108:3761–3766. doi: 10.1073/pnas.1014835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferlay J, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 11.Jung YJ, et al. Reciprocal expressions of cyclin E and cyclin D1 in hepatocellular carcinoma. Cancer Lett. 2001;168:57–63. doi: 10.1016/s0304-3835(01)00403-7. [DOI] [PubMed] [Google Scholar]
- 12.Peng SY, Chou SP, Hsu HC. Association of downregulation of cyclin D1 and of overexpression of cyclin E with p53 mutation, high tumor grade and poor prognosis in hepatocellular carcinoma. J Hepatol. 1998;29:281–289. doi: 10.1016/s0168-8278(98)80014-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Q, He Q, Liang LJ. Expression of p27, cyclin E and cyclin A in hepatocellular carcinoma and its clinical significance. World J Gastroenterol. 2003;9:2450–2454. doi: 10.3748/wjg.v9.i11.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 16.Sung WK, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 17.Cho JW, et al. Hepatitis C virus core protein promotes cell proliferation through the upregulation of cyclin E expression levels. Liver. 2001;21:137–142. doi: 10.1034/j.1600-0676.2001.021002137.x. [DOI] [PubMed] [Google Scholar]
- 18.Pok S, et al. Testosterone regulation of cyclin E kinase: A key factor in determining gender differences in hepatocarcinogenesis. J Gastroenterol Hepatol. 2016;31:1210–1219. doi: 10.1111/jgh.13232. [DOI] [PubMed] [Google Scholar]
- 19.Geng Y, et al. Cyclin E ablation in the mouse. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 20.Parisi T, et al. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J. 2003;22:4794–4803. doi: 10.1093/emboj/cdg482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Odajima J, et al. Cyclin E constrains Cdk5 activity to regulate synaptic plasticity and memory formation. Dev Cell. 2011;21:655–668. doi: 10.1016/j.devcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 23.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 24.Kalaszczynska I, et al. Cyclin A is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell. 2009;138:352–365. doi: 10.1016/j.cell.2009.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 26.Aleem E, Kiyokawa H, Kaldis P. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat Cell Biol. 2005;7:831–836. doi: 10.1038/ncb1284. [DOI] [PubMed] [Google Scholar]
- 27.Bishop AC, Shokat KM. Acquisition of inhibitor-sensitive protein kinases through protein design. Pharmacol Ther. 1999;82:337–346. doi: 10.1016/s0163-7258(98)00060-6. [DOI] [PubMed] [Google Scholar]
- 28.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 29.Zhang C, et al. A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat Methods. 2005;2:435–441. doi: 10.1038/nmeth764. [DOI] [PubMed] [Google Scholar]
- 30.Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 31.Puyol M, et al. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell. 2010;18:63–73. doi: 10.1016/j.ccr.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 32.Choi YJ, et al. The requirement for cyclin D function in tumor maintenance. Cancer Cell. 2012;22:438–451. doi: 10.1016/j.ccr.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov. 2016;6:353–367. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto Y, Maller JL. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science. 2004;306:885–888. doi: 10.1126/science.1103544. [DOI] [PubMed] [Google Scholar]
- 37.Geng Y, et al. Kinase-independent function of cyclin E. Mol Cell. 2007;25:127–139. doi: 10.1016/j.molcel.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 38.Santamaría D, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 39.Winter GE, et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geng Y, et al. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell. 1999;97:767–777. doi: 10.1016/s0092-8674(00)80788-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.