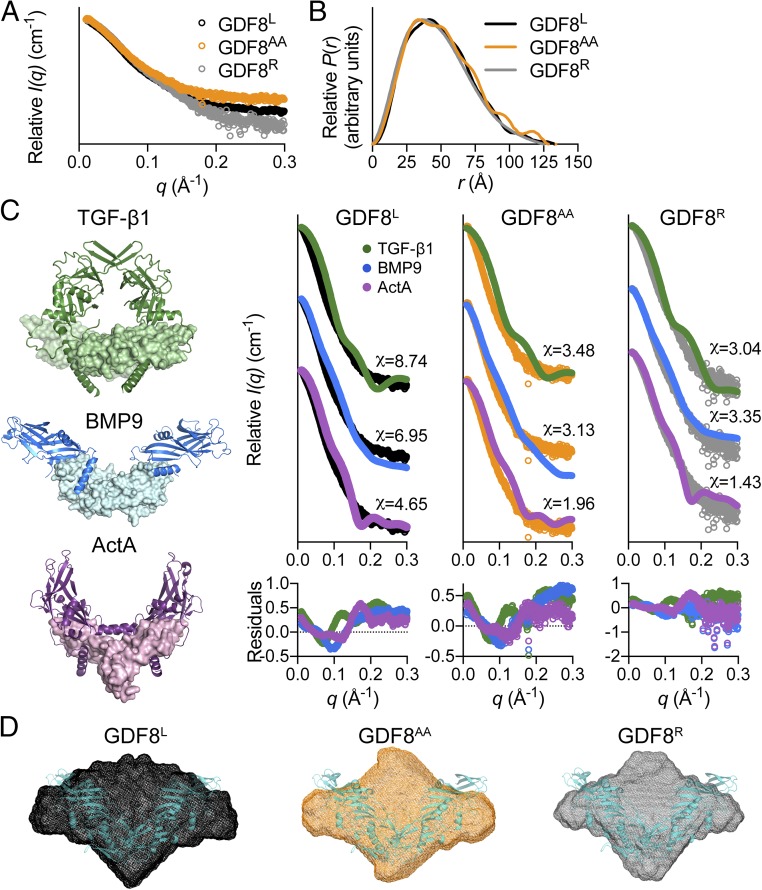
Fig. 2.
SAXS analysis of latent, acid-activated, and reformed GDF8 prodomain complex. (A) SAXS scattering profile showing the intensity distribution and (B) the pairwise distribution function for the various GDF8 prodomain complexes. (C) The crystal structures of various prodomain:ligand complexes used to generate theoretical scattering profiles for comparison [TGF-β1, Protein Data Bank (PDB) ID code 3RJR (18); BMP9, PDB ID code 4YCG (14); ActA (Activin A), PDB ID code 5HLY (15)]. The chi (χ) value was determined using the FoXS webserver (26). Residuals for each comparison are shown below the scattering profiles. Note that the latent TGF-β1 structure exemplifies a closed conformation unlike the nonlatent, but prodomain:ligand-associated BMP9 and Activin A (ActA) structures are in an open conformation. (D) Ab initio SAXS envelope (DAMFILT model) of the GDF8L (black), GDF8AA (orange), and GDF8R (gray) complexes. Note that the reconstruction of the GDF8AA complex appears more elongated compared with the other GDF8 prodomain complexes. The recently resolved GDF8 prodomain complex crystal structure [PDB ID code 5NTU (37)], shown in teal, is superimposed on the various ab initio molecular envelopes.