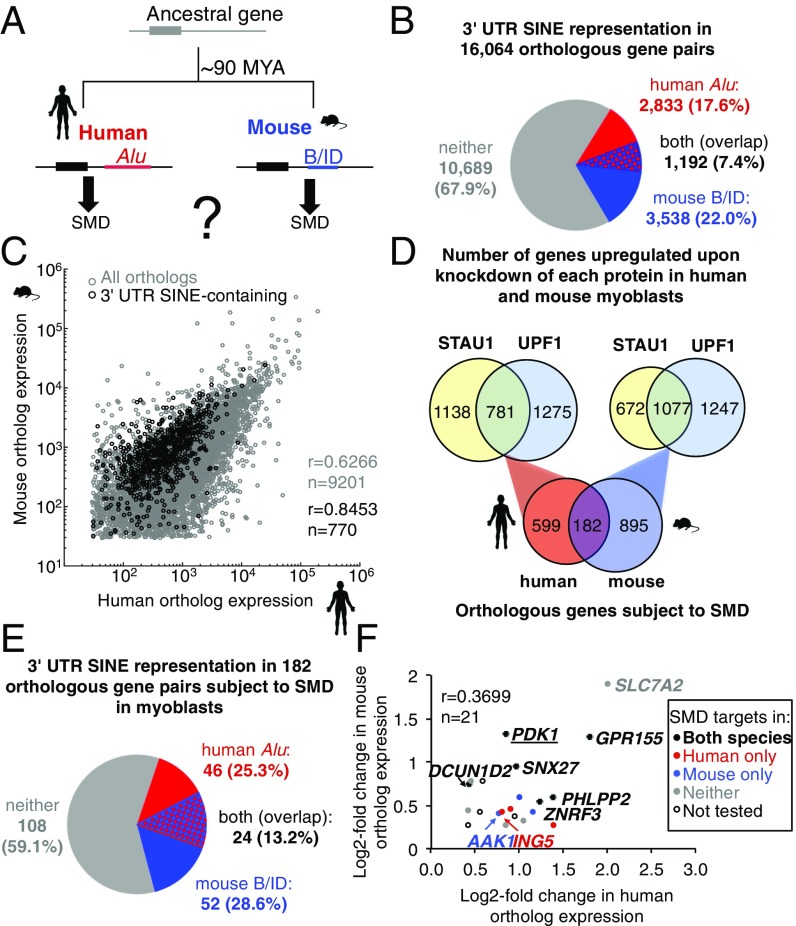
Fig. 1.
Identification of 3′-UTR SINE-containing orthopairs that are targets of Staufen-mediated mRNA decay (SMD). (A) Diagram of an archetypal orthopair that independently acquired a 3′-UTR Alu SINE in humans and a B or ID (B/ID) SINE in mouse. (B) Pie chart showing the proportion of human:mouse orthopairs that are 3′-UTR SINE-containing in human (red), mouse (blue), both (red and blue), or neither ortholog (gray). (C) Scatterplot showing the correlation in orthologous gene expression in control siRNA-treated human and mouse myoblasts (MBs); r, Pearson correlation coefficient; n, number of observations. (D) Venn diagrams demonstrate that hMBs and mMBs share 182 orthologs that are putative SMD targets, i.e., transcripts significantly up-regulated relative to their level in control siRNA-treated cells when STAU1 and, separately, UPF1 was depleted using siRNA in hMBs or mMBs. (E) Pie chart showing the proportion of the 182 orthopairs that produce putative SMD targets in both hMBs and mMBs and are 3′-UTR SINE-containing in the human ortholog (red), mouse ortholog (blue), or both orthologs (red and blue). (F) Scatterplot comparing the log2-fold change after hSTAU1 or mStau1 knockdown of each 3′-UTR SINE-containing human and mouse orthopair (3 of the 24 were excluded from this analysis as the major 3′-UTR isoform lacks the SINE). Color coding indicates SMD target validation using RT-qPCR (SI Appendix, Fig. S2).