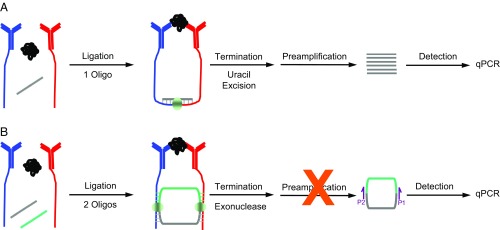
Fig. 1.
Schematic representation of t-PLA and c-PLA. (A) t-PLA detects proteins using pairs of antibody–DNA conjugates (red and blue), which are brought into close proximity on binding to target analyte. The addition of a bridge oligonucleotide and DNA ligase enables ligation of the antibody-tethered oligonucleotides to form a new DNA sequence. Ligation is terminated by selective degradation of the bridge oligonucleotide. The newly formed ligation product is subsequently preamplified followed by quantification using qPCR. (B) In c-PLA, the antibody-tethered oligonucleotides act as bridges for two ligation events between free oligonucleotides, resulting in the formation of a circular ligation product. The addition of an extra oligonucleotide increases stringency compared with t-PLA; it lowers the probability of random background ligation events, since four components must assemble in the absence of the target analyte to generate an independent circular ligation product. Circle formation also allows exonuclease treatment, which terminates ligation and reduces background by degrading all uncircularized DNA. The reduction in background also simplifies the workflow by eliminating the need for preamplification. Circular ligation products are quantified by qPCR using primer sites spanning the newly formed junctions (P1 and P2).