Significance
LPS is the major bacterial molecule recognized by the human innate immune system. It elicits a strong inflammatory response followed by a state of immune tolerance. The human enzyme acyloxyacyl hydrolase (AOAH) then detoxifies LPS to reestablish sensitivity for subsequent infections. We determined the 3D structure of AOAH to better understand its function. The enzyme binds to LPS via a hydrophobic surface and contains a hydrophobic tunnel into which one fatty acyl chain of the LPS fits to be cleaved off. AOAH also interacts with the phosphate groups of LPS but not its saccharide portion. Additionally, we report an unexpected calcium-binding domain in this enzyme.
Keywords: acyloxyacyl hydrolase, lipopolysaccharide, immune tolerance, saposin, calcium-binding protein
Abstract
LPS is a potent bacterial endotoxin that triggers the innate immune system. Proper recognition of LPS by pattern-recognition receptors requires a full complement of typically six acyl chains in the lipid portion. Acyloxyacyl hydrolase (AOAH) is a host enzyme that removes secondary (acyloxyacyl-linked) fatty acids from LPS, rendering it immunologically inert. This activity is critical for recovery from immune tolerance that follows Gram-negative infection. To understand the molecular mechanism of AOAH function, we determined its crystal structure and its complex with LPS. The substrate’s lipid moiety is accommodated in a large hydrophobic pocket formed by the saposin and catalytic domains with a secondary acyl chain inserted into a narrow lateral hydrophobic tunnel at the active site. The enzyme establishes dispensable contacts with the phosphate groups of LPS but does not interact with its oligosaccharide portion. Proteolytic processing allows movement of an amphipathic helix possibly involved in substrate access at membranes.
Lipopolysaccharide (LPS, endotoxin) is the major constituent of the external face of the Gram-negative bacterial outer membrane (1) and is the most potent agonist of innate immunity in humans (1). This complex molecule is composed of a glycolipid called “lipid A,” linked to a core oligosaccharide to which a long O-polysaccharide is attached (1, 2). Lipid A normally consists of a diphosphorylated diglucosamine scaffold bearing several lipid tails. Stimulation of the MD-2–Toll-like receptor 4 (TLR4) immune receptor complex by LPS at the cell surface requires the presence of six acyl chains on its lipid A moiety; penta- or hepta-acyl ligands are 100-fold less active, and tetra-acyl analogs are inactive (1–5). Hexa-acyl LPS is also necessary for the detection of bacterial invasion by cytoplasmic caspases 4, 5, and 11 (6–8).
Following the acute response, the host can enter a tolerant state in which subsequent responses to LPS are dampened (5, 9, 10), preventing excessive inflammation. Persistent exposure of macrophages to endotoxin down-regulates the expression of proinflammatory factors, including TNFα, and represses intracellular signal-transduction pathways initiated by TLR4 (11). This involves regulators, including SOCS1, ST2, IRAKM, SHIP, GILZ, and mir-146a, as well as transcriptional control (11). However, recovery from this state of immune tolerance is required for proper defense against further Gram-negative infections. Reestablishment of sensitivity to LPS is accomplished by its detoxification from the resolving infection (5, 9, 10, 12). This process is carried out by a host enzyme, acyloxyacyl hydrolase (AOAH), which removes fatty acids from lipid A (13, 14), thereby rendering it inert to TLR4- or caspase-based detection.
Lipid A typically contains four primary acyl chains directly attached to the diglucosamine backbone and a variable number of secondary fatty acids linked to the primary ones (Fig. 1C); AOAH selectively hydrolyzes the secondary (acyloxyacyl-linked) chains (13–19). This protein is found in endosomes or possibly lysosomes of phagocytic white blood cells (5, 13, 15, 18, 19) and liver-resident macrophages (20) as well as in the serum of certain animals (19, 21) and acts on LPS aggregates, bacterial membrane remnants, and possibly on LPS solubilized by the proteins CD14 or LBP (22). In mice, AOAH deficiency results in prolonged tolerance (over 2 months) (9), whereas its overexpression improves survival and recovery from bacterial infection (23). It has been suggested that enhancing AOAH activity could improve outcomes in infections and certain chronic conditions in humans (9, 23).
Fig. 1.
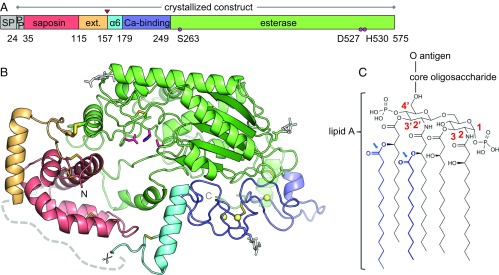
AOAH structure and substrate. (A) Domain organization of AOAH. The native proteolytic processing site is marked by a red triangle. Catalytic triad residues are represented by pink circles. α6, amphipathic helix; ext, saposin domain extension; PP, propeptide; SP, signal peptide. (B) Mature AOAH is composed of a small subunit containing a saposin domain (red) with an extension (orange) and a large subunit comprising an esterase domain (green), a calcium-binding region (dark blue), and an amphipathic helix α6 (cyan). The native proteolytic processing site is indicated by scissors, and a segment not visible in the structure is represented by a gray dashed line. The N and C termini are labeled (the propeptide is not visible in the structure). N-linked glycans (white sticks) are simplified for clarity. Calcium ions (yellow spheres) and disulfide bonds (yellow sticks) are displayed, including the interdomain bridge as thicker sticks. Catalytic triad residues are shown as pink sticks. (C) Chemical structure of E. coli LPS, with positions on the diglucosamine backbone of lipid A indicated in red. Secondary (acyloxyacyl) chains are in blue, with the bonds hydrolyzed by AOAH marked by blue arrows.
Although AOAH was discovered more than 30 years ago (13, 14), an understanding of the molecular details of AOAH function is still lacking. The determinants of LPS recognition by the protein are unknown, as is the basis of its selectivity toward secondary acyl chains. The specific role of its built-in saposin domain (19) is unclear, and the mechanism by which proteolytic processing of AOAH in acidic organelles increases its enzymatic activity on LPS (19) has not been determined. Here, we report the crystal structures of AOAH and its complex with lipopolysaccharide; these findings, together with biophysical and biochemical analyses, address these questions.
Results
Overall Structure.
Endosomal AOAH undergoes differential posttranslational processing relative to its secreted counterpart. The N-terminal signal peptide is removed from both forms, but a propeptide is further cleaved off only from the endosomal protein (17, 19). Additionally, a proteolytic event in the acidic environment of endosomes separates the enzyme into a small and a large subunit linked by a disulfide bridge (15, 17, 19). We determined the crystal structures of full-length human, murine, and rabbit AOAH lacking the signal peptide, including the structures of the intact and cleaved protein (Fig. 1 A and B). The large subunit comprises the catalytic domain, a hitherto unrecognized calcium-binding region, and an isolated amphipathic helix (α6). The small subunit consists of a saposin domain (17, 19); saposins are lipid- and membrane-binding modules that assist the function of lysosomal hydrolases, among other roles (24, 25). The propeptide is not visible in the electron density of the structures. The protein bears four Asn-linked glycans, whereas the Asn336 sequon is buried and inaccessible for glycosylation.
Catalytic and Saposin Domains.
The AOAH catalytic domain has a five-stranded parallel β-sheet at its core surrounded by 11 helices (Fig. 1B and Fig. S1A). This architecture is conserved in the GDSL esterases/lipases family (Pfam PF00657 and PF13472) (Fig. S1B) (17), of which the best-characterized mammalian member is the platelet-activating factor acetylhydrolase. The saposin domain is composed of four α-helices stabilized by three disulfide bonds and bears a C-terminal extension which contains the intersubunit disulfide bridge (Cys123 and Cys453) (Figs. 1B and 2A). The C-terminal portion of the extension is disordered in the structure and extends to the natural proteolytic processing site (Ser157). Saposins are conformationally flexible and adopt either a closed form that buries hydrophobic residues in the core or an open state with an exposed hydrophobic surface that participates in lipid and membrane binding (25). The saposin domain of AOAH is partially open, most closely resembling the conformation of saposin B or saposin C in the presence of detergent (Fig. 1B and Fig. S1C).
Fig. 2.
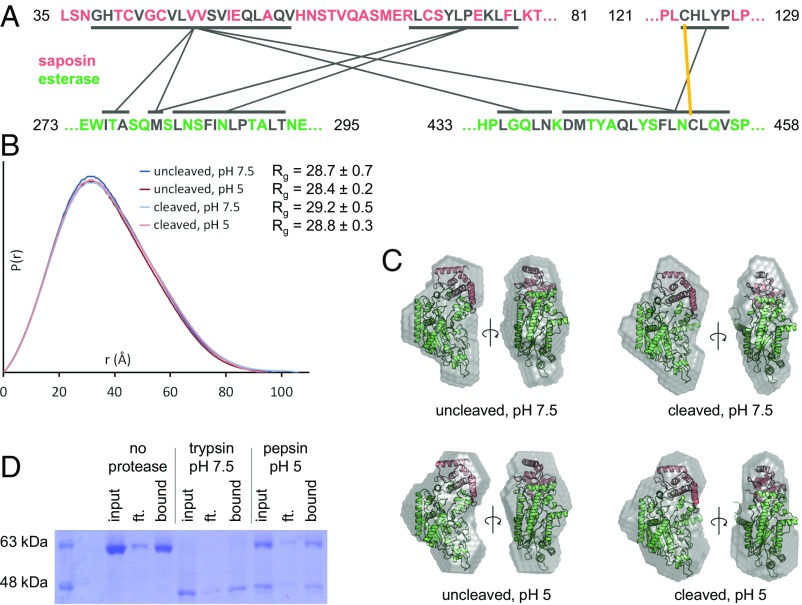
Position of the saposin domain relative to the esterase domain. (A) Residues forming the mainly hydrophobic interface between the saposin and esterase domains are in gray, with lines representing areas of contact. The interdomain disulfide bridge is indicated by a yellow line. (B) Distance distribution [P(r)] functions from SEC-SAXS experiments for uncleaved or cleaved human AOAH under different conditions. Rg, radius of gyration. (C) Shape reconstructions (gray) from SEC-SAXS experiments. The crystal structures (red and green) of uncleaved murine or cleaved human AOAH were automatically fitted into the SAXS envelopes. (D) To evaluate the interdomain interaction stability, human AOAH lacking the interdomain disulfide bridge (Cys123Ala mutant) and bearing a hexahistidine tag on the small subunit was incubated with or without protease at different pHs. Coprecipitation of the two-subunit complex (∼17 and 51 kDa) was then performed at pH 7.5 with Ni-NTA beads and analyzed on a nonreducing gel. Cleavage of AOAH by pepsin was incomplete. Ft, flowthrough.
Mammals possess four saposins (A–D) that assist lysosomal hydrolases in sphingolipid degradation, and saposins A and C form stable complexes with their cognate enzymes (24, 26). Moreover, in acid sphingomyelinase, the only mammalian protein besides AOAH that contains saposin and catalytic domains within a single polypeptide chain, the saposin domain can be in an open state and associate with the catalytic domain via a hydrophobic interface (27–29) or can adopt a closed conformation with fewer contacts to the rest of the enzyme (27). In AOAH, the relative position of the saposin domain is invariant in all the crystal structures reported here, including the intact and proteolytically processed forms, and in presence of substrates and detergent. The small subunit is abutted against two long segments of the catalytic domain (Fig. 1B), interacting with the latter via an extensive 1,165-Å2 hydrophobic interface involving 40 side chains, four hydrogen bonds, and a disulfide bridge (Fig. 2A).
Without the constraints of the crystal environment, however, motion of the saposin domain could be envisioned as occuring around the disulfide tether, particularly after proteolysis of the intersubunit linker. To detect such movements, the shape of the protein in solution was examined by small-angle X-ray scattering (SAXS) (Fig. S2). Proteolytic cleavage and changes in pH had no effect on the radius of gyration (the average distance of all atoms to the center) or the distance distribution function (the distribution of all interatomic distances) (Fig. 2B). Additionally, the reconstructed shape of the enzyme in solution was similar in all cases and agreed with the crystal structure (Fig. 2C). To verify the stability of the interdomain interface, the disulfide bond was mutated, and coprecipitation experiments were performed on proteolytically processed AOAH. Both subunits remained associated after 2 h at neutral and acidic pH (Fig. 2D). However, the total amount of recovered mutant protein was greatly decreased after proteolysis. Combined with the much lower expression yield of the disulfide mutant or Δsaposin construct, these observations suggest that slow dissociation of the subunits would destabilize the protein. In summary, the two subunits of AOAH form a rigid complex, although the possibility of rearrangement at membrane interfaces cannot be excluded.
Calcium-Binding Region.
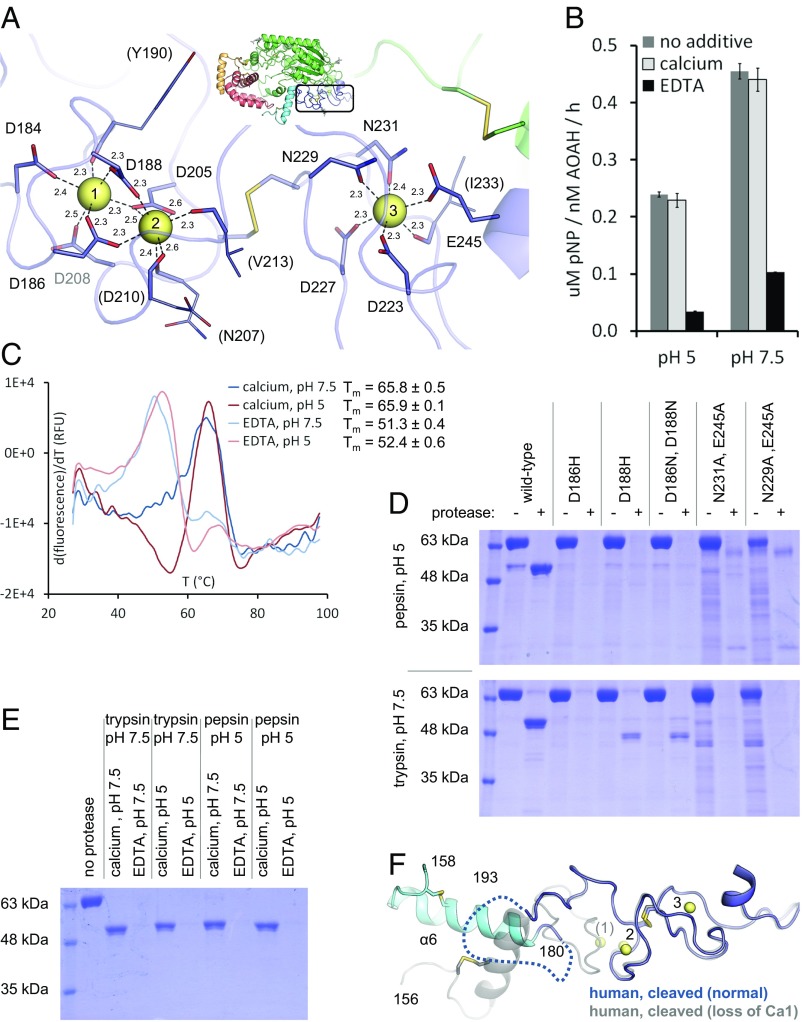
Electron density of the AOAH structures revealed the presence of metal ions in a region neighboring the catalytic domain (Fig. 1B). This 70-residue segment has no known sequence homology, including among GDSL esterases/lipases or saposins. Two adjacent metals are coordinated by five acidic side chains and four backbone carbonyl groups, and a third ion is held by three anionic residues, two asparagine side chains, and a backbone carbonyl group (Fig. 3A). Analysis of the crystallographic temperature factors and binding geometry indicated these metals are probably calcium (Fig. S3A). We sought to determine the purpose of this calcium-binding region in AOAH function. As discussed later, it is unlikely to interact with the LPS substrate (e.g., via its phosphate groups) since the calcium ions are fully coordinated by the protein. Moreover, the metal-binding residues are conserved even in AOAH homologs from unicellular organisms (Fig. S3B), some of which have substrates other than LPS (30). On the other hand, calcium may serve to stabilize the enzyme, as found in several other proteins (31). Indeed, prolonged incubation with EDTA decreased enzymatic activity on a small-molecule generic substrate (Fig. 3B) and lowered the melting temperature of AOAH in thermal stability assays (Fig. 3C). Additionally, EDTA treatment or mutation of calcium-binding residues rendered the enzyme highly susceptible to proteolytic degradation (Fig. 3 D and E). Last, the loss of one calcium ion was observed in one of the crystal structures, along with a local rearrangement and disordering (Fig. 3F). In summary, we identified a calcium-binding region that appears to be important for stability in AOAH.
Fig. 3.
Importance of the calcium-binding region for stability. (A) The calcium-binding region of AOAH (boxed area). Residues coordinating calcium ions (yellow spheres) are numbered, with numbers in parentheses for interactions via backbone carbonyl groups. Interatomic distances (in Ångstroms) are indicated. (B) To evaluate the importance of calcium for the protein’s integrity, human AOAH was first incubated with calcium or EDTA at pH 7.5, followed by an assay of activity against p-nitrophenyl acetate at different pHs. Data are the mean ± SD of three replicates representative of one of two experiments. pNP, p-nitrophenol. (C) Thermal stability of human AOAH in the presence of calcium or EDTA was measured using a hydrophobic fluorescent dye. One typical curve is displayed for each condition. Melting temperature (Tm) is the mean ± SD of four replicates. (D) Mutants of human AOAH that disrupt the coordination of calcium ions 1 and 2 (residues 186 and 188) or of the third calcium ion (residues 229, 231, 245) were incubated with or without protease at different pHs, and proteolytic stability was assessed on reducing gels. The lower yield of the last two mutants resulted in lower initial purity. (E) Wild-type human AOAH was first incubated with calcium or EDTA at different pHs, followed by protease treatment at different pHs and analysis on a reducing gel. (F) The structure of human AOAH (blue) that lost one calcium ion (yellow spheres) is compared with the protein (gray, transparent) with the full complement of three calcium ions. A segment not visible in the structure is represented by a dashed line.
LPS Recognition.
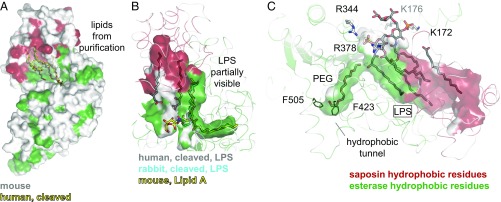
The surface of AOAH harbors a large, continuous hydrophobic pocket formed by the half-open saposin subunit together with adjacent portions of the catalytic domain. Structures of the enzyme crystallized in absence of substrates contained additional electron density within this cavity, attributed to fatty acids and phospholipids carried over from protein purification (Fig. 4A and Fig. S4). To verify whether the LPS substrate also binds at this site, catalytically inactive AOAH was crystallized in presence of Escherichia coli lipid A or LPS Ra mutant, bearing six acyl chains and the core oligosaccharide but lacking the O-antigen. In the resulting structures of the human, murine, and rabbit enzyme, the substrates are only partially visible in the electron density, with their fatty acid moieties bound in this pocket (Fig. 4B and Fig. S4). Notably, a single acyl chain often occupies a narrow lateral hydrophobic tunnel extending into the catalytic domain (Fig. 4B), likely a secondary (acyloxyacyl) chain based on connectivity. Furthermore, a structure of rabbit AOAH together with fully discernible lipid A was obtained (Fig. 4C and Fig. S4); however, a PEG molecule from crystallization fills the hydrophobic tunnel. The detailed interactions between LPS and the enzyme are addressed later; here the active site is described.
Fig. 4.
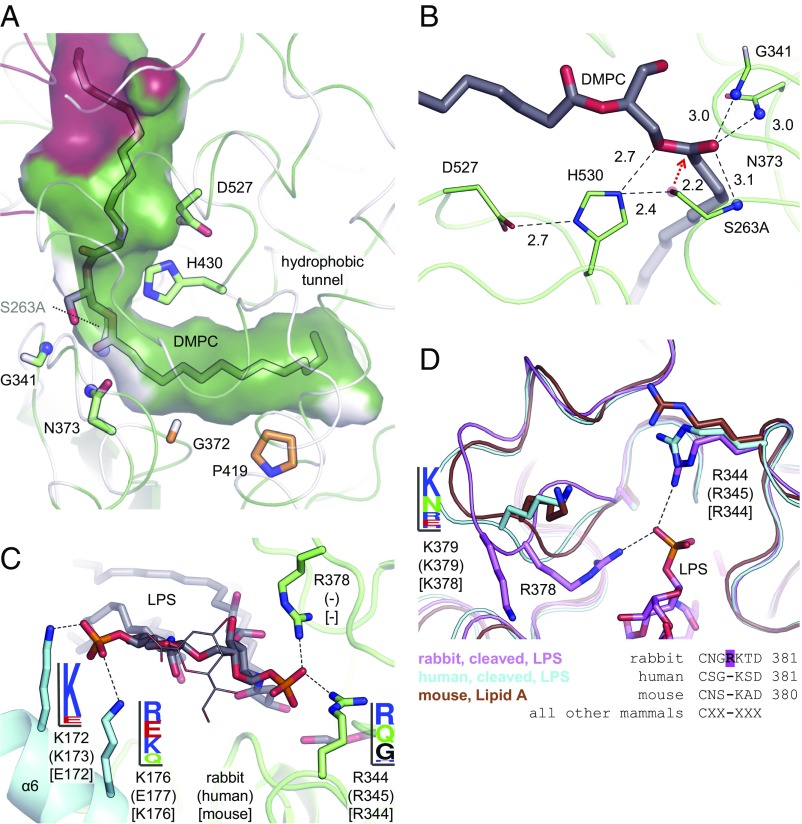
AOAH in complex with lipids and LPS. (A) Lipids carried over from purification and visible in the structure of human and murine AOAH are modeled as free fatty acids or phosphatidic acid (sticks). They are bound to a large hydrophobic pocket formed by the saposin (red) and esterase (green) domains. Helix α6 is omitted for clarity. (B) In complexes of the enzyme with LPS, a hydrophobic tunnel accommodates a secondary (acyloxyacyl) chain of the LPS ligands, which are only partially visible in the electron density. In A and B, the surface of the human protein is displayed. (C) In a mostly complete structure of LPS bound to rabbit AOAH, a PEG molecule occupies the hydrophobic tunnel. One acyl chain of the lipid A moiety is not clearly visible in the electron density. Cationic side chains that form salt bridges with the phosphate groups of LPS, as well as two phenylalanine residues capping the hydrophobic tunnel, are displayed as sticks and numbered.
The lateral hydrophobic tunnel entrance is located next to a set of residues conserved in the GDSL esterases/lipases family (Fig. S5) that form the active site. To gain a clearer view of the catalytic mechanism, a high-resolution structure of inactive murine AOAH in complex with phosphatidylcholine was determined; phospholipids can also serve as substrates for this enzyme (18, 19). The lipid is bound with one acyl chain inserted into the tunnel, whereas the phosphocholine head group is not discernible (Fig. 5A and Fig. S4). The nucleophilic serine hydroxy group (Ser263Ala mutant) is positioned near the carboxyl carbon of that acyl chain (Fig. 5B). Neighboring His530 and Asp527 complete the catalytic triad. The reaction mechanism of GDSL esterases/lipases begins with a nucleophilic attack by the serine side chain on the substrate’s ester carbon, with the oxyanion intermediate stabilized by the backbone amide groups of Gly341 and Ser263 and the side chain of Asn373, which together form the oxyanion hole (Fig. 5B) (32). Release of the substrate (here, deacylated LPS) results in an acyl-enzyme covalent intermediate. AOAH can transfer fatty acids between lipids (18) via this intermediate. Subsequent hydrolysis dissociates the acyl chain and regenerates the active site.
Fig. 5.
Active site and LPS phosphate recognition. (A and B) In the structure of murine AOAH in complex with DMPC, one acyl chain is inserted into the hydrophobic tunnel. The phosphocholine moiety is not visible in the electron density. Residues involved in catalysis are displayed as green sticks, with the inactivating mutation Ser263Ala. Two amino acids lining the hydrophobic tunnel are in orange. Hydrogen bonds and salt bridges are represented by dashed lines. The Ser263 oxygen atom is modeled as a transparent red sphere with a red arrow representing the nucleophilic attack. The amide groups forming the oxyanion hole are shown as blue spheres. Interatomic distances (in Ångstroms) are indicated. Residues are numbered according to the human protein. (C) Cationic residues in rabbit AOAH that establish salt bridges (dashed lines) with the phosphate groups of LPS (sticks) are displayed as sticks. Their conservation in vertebrates is depicted as sequence logos. For C and D, rabbit, human (in parentheses) and murine (in square brackets) residues are numbered. (D) A loop contains the rabbit-specific Arg378 that forms a salt bridge (dashed line) with a phosphate group of LPS. An adjacent lysine residue is also displayed, with its conservation in vertebrates depicted as a sequence logo. The sequence of this loop in mammals is aligned.
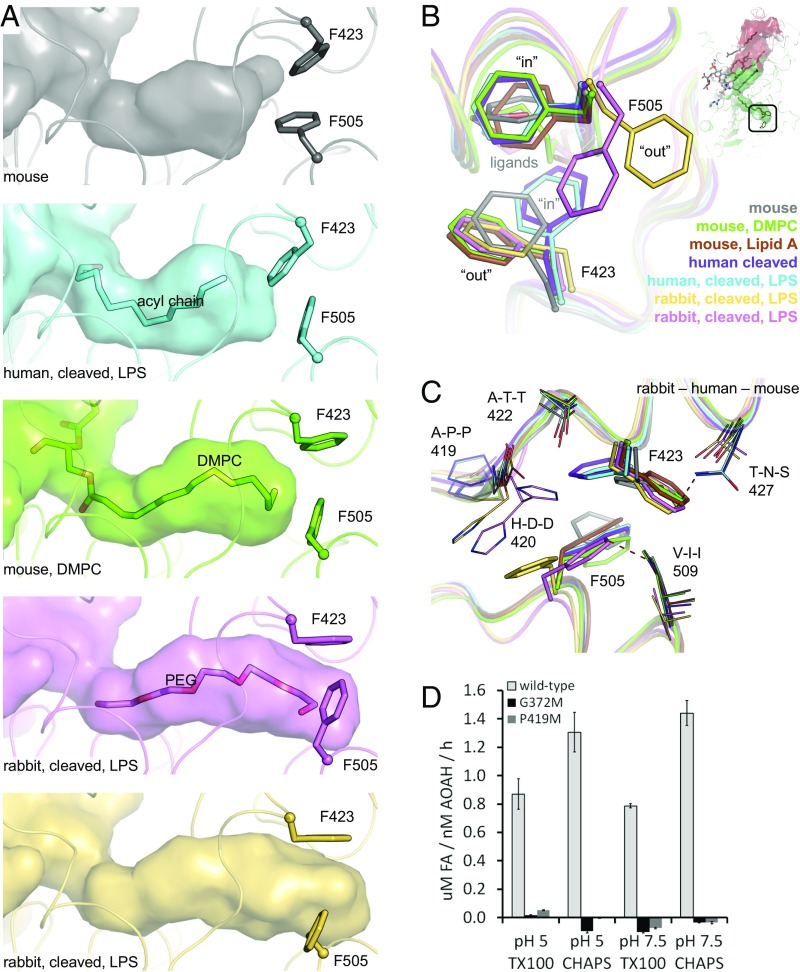
To validate the hydrophobic tunnel as the binding site of the secondary (acyloxyacyl) chains of LPS, two residues lining its walls (Gly372 and Pro419) (Fig. 5A) were replaced by methionine to block lipid entry. The mutations did not negatively affect the fold of the protein as judged by enzymatic activity on a small-molecule generic substrate (Fig. S6A). However, deacylation of LPS was abolished (Fig. 6D), confirming the hydrophobic tunnel’s role. The further end of this narrow cavity is capped by the side chains of Phe423 and Phe505 (Fig. 4C). Interestingly, the depth of the tunnel varies between the structures reported here, being restricted by differing orientations of the two phenylalanine side chains (Fig. 6A) which adopt either “in” or “out” conformations (Fig. 6B). This variability does not seem to be induced by the presence or absence of substrate. Rather, species-specific differences in the amino acids surrounding the phenylalanine residues could favor certain orientations (Fig. 6C), with the rabbit enzyme generally displaying a deeper cavity.
Fig. 6.
Variability of the hydrophobic tunnel. (A) The depth of the hydrophobic tunnel in different AOAH structures is compared. (B) Side-chain orientations of the two phenylalanine residues capping the hydrophobic tunnel are compared for different AOAH structures, with positions labeled as “in” (smaller tunnel) or “out” (larger tunnel). (C) Species-specific local variations that could contribute to the different orientations of tunnel-capping phenylalanine residues (thick sticks) are displayed as thin sticks. Residue identity at each position is for rabbit–human–mouse. Two conformations are visible for His420. Potential clashes across species are represented by red dashed lines. (D) To validate the role of the hydrophobic tunnel, mutations were introduced to block the tunnel (see also Fig. 5A), and the activity of human AOAH against LPS in Triton X-100 or CHAPS micelles was assayed at different pHs. Data are the mean ± SD of three replicates representative of one of two experiments. For the entire figure, residues are numbered according to the human protein. FA, free fatty acids.
AOAH can remove secondary fatty acids at positions 3′, 2′, and 2 of the lipid A diglucosamine backbone (16). LPS from certain bacteria also contains an acyloxyacyl moiety at position 3 (2) and perhaps could serve as substrate as well, although this has not been verified. This wide specificity is made possible by the ability of the enzyme to bind to lipid A in different orientations. In several structures reported here, LPS is only partially distinguishable, likely due to multiple substrate conformations in the crystal. Nevertheless, the primary acyl chain that bears the secondary fatty acid to be cleaved is clearly visible in all cases (Fig. 4B), providing part of the explanation for the protein’s selectivity for secondary acyl chain hydrolysis; the two hydrocarbon tails of the acyloxyacyl moiety together appear to be major contributors to binding affinity, masking a continuous hydrophobic surface on the enzyme. Furthermore, complete insertion of a primary acyl chain into the hydrophobic tunnel, required to position the ester bond at the active site, would be sterically hindered by the substituents on the glucosamine, while entry of secondary fatty acids allows more distance to the bulky saccharide. The tightness and capping of the tunnel also account for the protein’s preference for nonhydroxylated, saturated, and short acyl chains (16, 18).
In summary, LPS binds to AOAH with its fatty acid tails covered by the hydrophobic pocket formed by the saposin and catalytic domains and a secondary (acyloxyacyl) chain buried in the hydrophobic tunnel at the active site. Notably, the protein does not interact with the diglucosamine backbone of lipid A or the core oligosaccharide, which is mostly disordered in the structures, and is not expected to bind the O-antigen, either. On the other hand, electrostatic contacts are established between the lipid A phosphate moieties and two sets of cationic residues (Figs. 4C and 5C), one located on loops from the catalytic domain and the other on amphipathic helix α6 (discussed later). These residues are moderately conserved in vertebrates (Fig. 5C). Surprisingly, Arg378 is unique to rabbits among all mammals (Fig. 5D). Mutation of these basic residues in the human enzyme did not affect LPS hydrolysis in our detergent-based in vitro assay (Fig. S6B). Moreover, deacylation at positions 2′ and 3 (Fig. 1C) entails LPS binding in a manner likely to disrupt contacts with one or both phosphates, as opposed to our structure which is representative of the binding mode for positions 3′ or 2 (Figs. 1C and 4C). However, these reactions occur at similar rates (16). Moreover, the phosphate moieties are substituted in certain bacteria, decreasing or neutralizing their charge (2); AOAH still remains active on one such substrate in vitro (16). Combined, these results suggest that electrostatic interactions are not crucial for LPS recognition, although a possible relevance in vivo cannot be excluded, especially in the rabbit ortholog.
Amphipathic Helix.
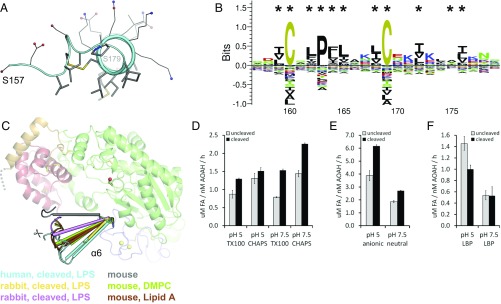
As mentioned previously, endosomal proteolysis separates AOAH into two subunits, but no major rearrangements were discerned in the crystal structures or by SAXS. The only variability occurs in helix α6 situated at the N terminus of the large subunit, immediately following the processing site. This helix adopts a range of orientations in both the intact and cleaved structures (Fig. 7C), with Ser179 as the hinge point, and wider rotation could be envisioned in the processed enzyme outside the crystal constraints. The small subunit also contains a C-terminal linker that becomes flexible after proteolytic cleavage (Fig. 1B); however its sequence conservation is low, and its presence in vivo is uncertain (17, 19).
Fig. 7.
Amphipathic helix of AOAH. (A) A view along the axis of the human AOAH amphipathic helix α6, with hydrophobic residues located on its nonpolar side displayed as thick sticks. The N terminus of the proteolytically processed large subunit (Ser157) is indicated, as is the point of attachment to the calcium-binding region (Ser179). (B) Conservation of residue class in the vertebrate AOAH amphipathic helix is depicted as a sequence logo, with hydrophobic residues (black) located on the nonpolar side marked by asterisks. (C) The orientation of the amphipathic helix α6 in different AOAH structures is compared. In cases with two protein chains per asymmetric unit, both are displayed. The native proteolytic processing site is indicated by scissors, and a segment not visible in the structure is represented by a gray dashed line. The active site Ser263 is shown as sticks and a red sphere. (D–F) The effect of proteolytic processing on the enzymatic activity of human AOAH was assessed at different pHs with LPS in Triton X-100 or CHAPS micelles (D), lipid A in anionic or neutral liposomes (E), or LPS assisted by small amounts of LBP (F). Data are the mean ± SD of three replicates representative of one of two experiments. FA, free fatty acids.
Proteolytic processing of AOAH greatly increases the in vitro hydrolysis rate of LPS, but not that of phospholipids, by an unknown mechanism (19). As no other structural changes were evident upon proteolysis, helix α6 was examined more closely. This segment is amphipathic, with one side partly facing the protein and bearing only hydrophobic residues, whereas the other side is solvent-exposed and displays only charged and polar residues (Fig. 7A and Fig. S7A). The hydrophobic portions of the helix are conserved in vertebrates (Fig. 7B). We initially evaluated the effect of cleavage on catalytic competency. Helix α6 is located far from the active site (Fig. 7C), and proteolysis did not enhance activity against a small-molecule generic substrate (Fig. S7B); in fact, a slight decrease was observed, possibly due to lowered stability of the protein. In contrast, processing increased the deacylation of LPS in a detergent-based assay up to twofold (Fig. 7D). A similar effect was measured with liposomes containing lipid A as substrate (Fig. 7E).
These results suggest that proteolysis could somehow improve LPS binding to the enzyme. However, the hydrophobic face of α6 does not interact with LPS acyl chains, at least in their final bound position (Fig. 8C). The electrostatic contacts between α6 and the phosphate group of lipid A are also not essential for activity, as stated before. Alternately, the amphipathic helix could facilitate access to the substrate in a micellar or membrane setting. It has been suggested that, in addition to LPS aggregates, bacterial fragments, or lipoprotein particles, AOAH can act on lipopolysaccharide extracted by the cofactor LPS-binding protein (LBP) (22). Remarkably, proteolytic processing did not increase LBP-assisted LPS deacylation and even decreased it at acidic pH (Fig. 7F). Processing thus could affect membrane-interacting properties of the enzyme. An intriguing observation is the ability of AOAH to bind to intact bacteria, but only at acidic pH and in its uncleaved form (Fig. 8E and Fig. S8B). Finally, these effects are not restricted to LPS. We demonstrate that the enzyme can also deacylate in vitro synthetic lipopeptides that mimic the lipoprotein ligands of TLR2 (33); processing also increases activity on these substrates in detergent micelles (Fig. 8D). In summary, endosomal proteolysis of AOAH may modulate access to substrates in a membrane environment, possibly via the amphipathic helix α6.
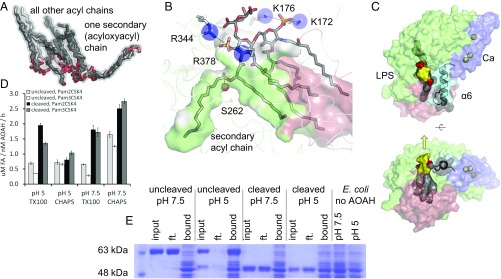
Fig. 8.
AOAH binding to LPS and other substrates. (A) The lipid moieties of LPS and related ligands from all AOAH structures are superimposed, illustrating a common set of hydrophobic tracks that accommodate the acyl chains. The chain to be hydrolyzed occupies the hydrophobic tunnel. (B) LPS bound in a productive orientation to rabbit AOAH is manually modeled based on the structures of complexes reported here. Electrostatic interactions are indicated by dashed lines. The catalytic serine oxygen atom is represented by a red sphere. (C) Relative positions of LPS, the amphipathic helix (α6), and the calcium-binding region (Ca) are illustrated. LPS is displayed with acyl chains (gray), the carbohydrate moiety (yellow), and phosphate groups (red). The amphipathic helix (α6, cyan) has its hydrophobic face in gray. Calcium ions are represented by yellow spheres. A yellow arrow indicates the expected direction of the core oligosaccharide extension. (D) The activity of processed or unprocessed human AOAH against the synthetic lipopeptides Pam2CSK4 and Pam3CKS4 in Triton X-100 or CHAPS micelles was assayed at different pHs. Data are the mean ± SD of three replicates representative of one of two experiments. FA, free fatty acids. (E) To test the binding of human AOAH to intact E. coli, the enzyme was incubated with bacteria at different pHs, centrifuged at low speed, and visualized on a reducing gel. The 53-kDa band in the uncleaved pH 5 input sample is a degradation product arising from a protease trace impurity. Ft, flowthrough.
Discussion
Detoxification of LPS is an essential step in recovery from Gram-negative bacterial infections, and the structures presented here reveal the molecular details of AOAH function. The protein removes secondary acyl chains from LPS, rendering this potent immunostimulatory agent inert to TLR- and caspase-based detection systems. The multiple structures of the enzyme in complex with LPS and other lipids enable us to propose a model of productive LPS binding. Acyl chains of lipid ligands are accommodated in a set of hydrophobic tracks formed by the saposin and catalytic domains, with one tail buried in a lateral hydrophobic tunnel for hydrolysis (Fig. 8A). Cleavage of the remaining secondary acyl chain(s) would therefore require at least partial dissociation and rebinding of the substrate. The LPS molecule bound to AOAH resembles an open palm, with the fingers representing primary lipid tails and the thumb designating a secondary acyl chain (Fig. 8B). The enzyme covers its substrate like a mitten, without contacting the core oligosaccharide or O-antigen (arm). Interestingly, the features recognized by AOAH, namely the lipid portion and phosphate groups, are also the determinants of LPS binding to and activation of the MD-2–TLR4 complex (Fig. S8A).
Polymorphisms in AOAH have been associated with asthma (34) and chronic rhinosinusitis (35). Over 200 single-nucleotide variants in this enzyme have also been identified from genome-sequencing projects. Based on the crystal structure as well as sequence conservation, we assessed the expected impact of each mutation on the protein’s stability, fold, or activity (Table S2). Whereas most polymorphisms are predicted to be benign, about 30 variants are likely harmful. AOAH participates in recovery from Gram-negative infections; thus, in addition to shedding light on the molecular mechanism of AOAH, our results could help identify individuals with impaired immune function.
Materials and Methods
Protein Expression and Purification.
Recombinant AOAH was expressed as a secreted protein in Sf9 insect cells infected with baculovirus. The endogenous signal peptide comprising the first 22 or 23 residues was replaced by the melittin signal peptide MKFLVNVALVFMVVYISYIYA followed by a hexahistidine tag DRHHHHHHKL. Constructs encompassed residues 24–575 of human (UniProt: P28039), residues 23–574 of murine (UniProt: O35298), and residues 23–575 of rabbit (UniProt: O18823) AOAH. All mutants were verified by DNA sequencing. AOAH was isolated from expression culture medium using nickel-nitrilotriacetic acid (Ni-NTA) resin (Thermo Fisher Scientific), further purified by size-exclusion chromatography on a Superdex 200 column (GE Healthcare) in buffer [15 mM Tris⋅HCl (pH 7.5), 100 mM NaCl], applied to a Q Sepharose anion exchange column (GE Healthcare), collected in the flowthrough, and concentrated to 10 mg/mL.
Crystallization.
For proteolytic processing for crystallization, human AOAH at 10 mg/mL was incubated with 10 μg/mL chymotrypsin at 4 °C for 16 h, while 33 μg/mL trypsin was used for the rabbit ortholog. Proteolysis was stopped with 0.5 mM PMSF. For complexes with lipids or LPS, inactive AOAH (Ser263Ala mutant) was incubated with 1 mM Triton X-100 and substrates: 0.333 mM E. coli LPS Ra (Sigma L9641), 1 mM E. coli lipid A (Sigma L5399), or 2 mM dimyristoyl phosphatidylcholine (DMPC) (Avanti 850345). Crystals were grown by sitting- or hanging-drop vapor diffusion at 22 °C and were flash-frozen after a brief soaking in a well solution supplemented with 20% glycerol. Well solutions were as follows: human cleaved [100 mM sodium citrate (pH 5) with 20% PEG 6000]; human cleaved with LPS [200 mM ammonium chloride with 100 mM sodium acetate (pH 5.3) and 20% PEG 6000]; rabbit cleaved with LPS structure with low-quality saposin [1 M sodium citrate (pH 7)]; rabbit cleaved with LPS [100 mM sodium MES (pH 6) with 34% PEG 200 and 5% PEG 3000 (15% PEG 200 was used as cryoprotectant instead of glycerol)]; murine [100 mM sodium acetate (pH 4.6) with 15% PEG 20000]; murine with lipid A or DMPC [100 mM sodium MES (pH 6) with 15% PEG 20000]. X-ray diffraction data were collected at 100 K on beamline 08ID-1 with a Rayonix MX300 CCD detector at the Canadian Macromolecular Crystallography Facility, Canadian Light Source. Data were processed by HKL2000 (36) with auto-corrections enabled or XDS (37).
Structure Determination.
A crystal of the cleaved rabbit AOAH with LPS (structure with low-quality saposin) was briefly soaked in well solution supplemented with 1 M sodium iodide, and the structure was solved by iodine single-wavelength anomalous diffraction using Autosol (38) in Phenix (39) and was manually built in Coot (40). The saposin domain could not be built initially due to excessive positional flexibility in this crystal form; the structure was completed by molecular replacement with the other crystal forms using Phaser (41) in Phenix. Refinement was carried out by phenix.refine (42). Translation–libration–screw (TLS) parameters were applied for the rabbit AOAH with LPS (structure with low-quality saposin) and the murine enzyme apo form and with DMPC. Noncrystallographic symmetry restraints were applied to the murine AOAH with lipid A. Crystallographic data collection and structure refinement statistics are presented in Table S1. Structural images were prepared with the PyMOL Molecular Graphics System, version 1.3 (Schrödinger, LLC). Sequence logos were created by the Seq2Logo server (43) with clustering disabled (Fig. 5) or enabled (Fig. 7).
SAXS.
Human AOAH was optionally proteolytically processed by chymotrypsin as described above. Size-exclusion chromatography-coupled SAXS (SEC-SAXS) experiments were carried out at the G1 beamline with a Dectris Pilatus 100K-S detector at the BioSAXS facility, Macromolecular Diffraction Facility of the Cornell High Energy Synchrotron Source (MacCHESS). Data were processed by BioXTAS RAW (44). Radii of gyration were calculated by PRIMUS (45) and distance distribution curves by GNOM (46) as part of ATSAS (47). Shape reconstructions were performed by DAMMIF (48) and crystal structures were fit into the SAXS envelopes by SUPCOMB (49).
Coprecipitation.
To evaluate the stability of the interaction between the small and large subunits, human AOAH lacking the interdomain disulfide bridge (Cys123Ala mutant) at 0.45 mg/mL was incubated at 4 °C for 2 h in buffer (100 mM NaCl, 5 mM CaCl2) with either 10 μg/mL trypsin and 10 mM Tris⋅HCl (pH 7.5) or 1 mg/mL pepsin and 10 mM sodium acetate (pH 5). Tris⋅HCl (pH 7.5) was then added to a final concentration of 100 mM along with Ni-NTA beads, followed by incubation at 4 °C for 1 h. Binding of human, murine, or rabbit AOAH to intact bacteria was assayed by incubating the protein at 0.1 mg/mL, either uncleaved or after tryptic processing, with E. coli at an optical density of 7, washing in binding buffer [100 mM NaCl, 50 mM Tris⋅HCl (pH 7.5) or sodium acetate (pH 5)] for 15 min at 22 °C, and centrifugation at low speed.
Stability Analysis.
Thermal stability of human AOAH at 4 μM was assayed in buffer [100 mM NaCl, 50 mM Tris⋅HCl (pH 7.5) or sodium acetate (pH 5) with 5 mM CaCl2 or EDTA] in the presence of 1× Protein Thermal Shift dye (Thermo Fisher Scientific). Melt curve experiments were performed using recommended settings. Melting temperatures were calculated by the derivative method with the Protein Thermal Shift Software. Proteolytic stability was assessed by incubating human AOAH mutants at 0.2 mg/mL for 2 h at 37 °C in buffer (100 mM NaCl, 5 mM CaCl2) with either 10 μg/mL trypsin and 50 mM Tris⋅HCl (pH 7.5) or 0.1 mg/mL pepsin and 50 mM sodium acetate (pH 5). Alternatively, the wild-type protein at 0.2 mg/mL was incubated at 37 °C for 24 h in buffer [100 mM NaCl, 20 mM Tris⋅HCl (pH 7.5) or sodium acetate (pH 5) with 5 mM CaCl2 or EDTA] followed by the addition of either 100 mM Tris·HCl (pH 7.5) with 10 μg/mL trypsin, or 100 mM sodium acetate (pH 5) with 0.1 mg/mL pepsin, and incubation at 37 °C for 2 h. To determine the effect of calcium removal on enzymatic activity, human AOAH was incubated at 37 °C for 24 h in buffer [100 mM NaCl, 10 mM Tris⋅HCl (pH 7.5) with 5 mM CaCl2 or EDTA] and diluted fivefold with enzymatic assay buffers (see below).
Enzymatic Activity.
Hydrolysis of p-nitrophenyl acetate was assayed at 2 mM substrate and 2 μM AOAH in buffer [100 mM NaCl, 20 mM Tris⋅HCl (pH 7.5) or sodium acetate (pH 5)] at 37 °C for 30 min. Absorbance was then measured at 405 nm for pH 7.5 samples or at 340 nm for pH 5 samples, and product formation was quantified with p-nitrophenol standard curves. LPS deacylation was measured by the Free Fatty Acid Fluorometric Assay (Cayman Chemical). AOAH at 0.1–0.25 μM was incubated with 0.25 mM E. coli LPS Ra (Sigma L9641) and detergent (1 mM Triton X-100 or 10 mM CHAPS) in buffer [100 mM NaCl, 20 mM Tris⋅HCl (pH 7.5) or sodium acetate (pH 5)] at 37 °C for 1 h. The reaction was stopped at 95°C for 5 min, and free fatty acids were quantified with a myristic acid standard curve. Deacylation of synthetic lipopeptides was also assayed in presence of detergent, with 0.5 mM Pam3CSK or Pam2CKS4 (InvivoGen). Lipid A liposomes were prepared by extrusion through 100-nm polycarbonate filters. Neutral liposomes consisted of 70 mol % phosphatidylcholine, 20% cholesterol and 10% E. coli lipid A (Sigma L5399); in anionic liposomes, 20% phosphatidylcholine was replaced by bis(monoacylglycero)phosphate. For assays with LBP, the LBP-assisted hydrolysis rate was obtained by subtracting activity against 0.25 mM LPS alone from activity in the presence of 10 μM LBP and 0.25 mM LPS.
Data Availability.
Atomic coordinates and structure factors were deposited into the Protein Data Bank (PDB) under the ID codes 5W78, 5W7C, 5W7A, 5W7B, 5W7D, 5W7E, and 5W7F.
Supplementary Material
Acknowledgments
We thank Dr. Robert Munford for providing the murine and rabbit AOAH cDNAs, helpful discussions, and advice and Dr. Shaun Labiuk for crystallographic data collection. The research described in this paper was performed using beamline 08ID-1 at the Canadian Light Source, which is supported by the Canada Foundation for Innovation, Natural Sciences and Engineering Research Council of Canada, the University of Saskatchewan, the Government of Saskatchewan, Western Economic Diversification Canada, the National Research Council Canada, and the Canadian Institutes of Health Research (CHIR). This work is based on research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation (NSF) and the NIH/National Institute of General Medical Sciences (NIGMS) under NSF Award DMR-0936384, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by Award GM-103485 from the NIH through the NIGMS. B.N. is supported by CIHR Operating Grant MOP-133535.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB), www.wwpdb.org (PDB ID codes 5W78 and 5W7A–5W7F).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719834115/-/DCSupplemental.
References
- 1.Molinaro A, et al. Chemistry of lipid A: At the heart of innate immunity. Chemistry. 2015;21:500–519. doi: 10.1002/chem.201403923. [DOI] [PubMed] [Google Scholar]
- 2.Kabanov DS, Prokhorenko IR. Structural analysis of lipopolysaccharides from gram-negative bacteria. Biochemistry (Mosc) 2010;75:383–404. doi: 10.1134/s0006297910040012. [DOI] [PubMed] [Google Scholar]
- 3.Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 4.Kim HM, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Munford R, Lu M, Varley A. Chapter 2: Kill the bacteria...and also their messengers? Adv Immunol. 2009;103:29–48. doi: 10.1016/S0065-2776(09)03002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kayagaki N, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 7.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: Implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi J, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 9.Lu M, Varley AW, Ohta S, Hardwick J, Munford RS. Host inactivation of bacterial lipopolysaccharide prevents prolonged tolerance following gram-negative bacterial infection. Cell Host Microbe. 2008;4:293–302. doi: 10.1016/j.chom.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu M, Varley AW, Munford RS. Persistently active microbial molecules prolong innate immune tolerance in vivo. PLoS Pathog. 2013;9:e1003339. doi: 10.1371/journal.ppat.1003339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins PE, Carmody RJ. The regulation of endotoxin tolerance and its impact on macrophage activation. Crit Rev Immunol. 2015;35:293–323. doi: 10.1615/critrevimmunol.2015015495. [DOI] [PubMed] [Google Scholar]
- 12.Zou B, et al. Acyloxyacyl hydrolase promotes the resolution of lipopolysaccharide-induced acute lung injury. PLoS Pathog. 2017;13:e1006436. doi: 10.1371/journal.ppat.1006436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall CL, Munford RS. Enzymatic deacylation of the lipid A moiety of Salmonella typhimurium lipopolysaccharides by human neutrophils. Proc Natl Acad Sci USA. 1983;80:6671–6675. doi: 10.1073/pnas.80.21.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munford RS, Hall CL. Detoxification of bacterial lipopolysaccharides (endotoxins) by a human neutrophil enzyme. Science. 1986;234:203–205. doi: 10.1126/science.3529396. [DOI] [PubMed] [Google Scholar]
- 15.Munford RS, Hall CL. Purification of acyloxyacyl hydrolase, a leaukocyte enzyme that removes secondary acyl chains from bacterial lipopolysaccharides. J Biol Chem. 1989;264:15613–15619. [PubMed] [Google Scholar]
- 16.Erwin AL, Munford RS. Deacylation of structurally diverse lipopolysaccharides by human acyloxyacyl hydrolase. J Biol Chem. 1990;265:16444–16449. [PubMed] [Google Scholar]
- 17.Hagen FS, et al. Expression and characterization of recombinant human acyloxyacyl hydrolase, a leukocyte enzyme that deacylates bacterial lipopolysaccharides. Biochemistry. 1991;30:8415–8423. doi: 10.1021/bi00098a020. [DOI] [PubMed] [Google Scholar]
- 18.Munford RS, Hunter JP. Acyloxyacyl hydrolase, a leukocyte enzyme that deacylates bacterial lipopolysaccharides, has phospholipase, lysophospholipase, diacylglycerollipase, and acyltransferase activities in vitro. J Biol Chem. 1992;267:10116–10121. [PubMed] [Google Scholar]
- 19.Staab JF, Ginkel DL, Rosenberg GB, Munford RS. A saposin-like domain influences the intracellular localization, stability, and catalytic activity of human acyloxyacyl hydrolase. J Biol Chem. 1994;269:23736–23742. [PubMed] [Google Scholar]
- 20.Shao B, et al. A host lipase detoxifies bacterial lipopolysaccharides in the liver and spleen. J Biol Chem. 2007;282:13726–13735. doi: 10.1074/jbc.M609462200. [DOI] [PubMed] [Google Scholar]
- 21.Erwin AL, Munford RS. Plasma lipopolysaccharide-deacylating activity (acyloxyacyl hydrolase) increases after lipopolysaccharide administration to rabbits. Lab Invest. 1991;65:138–144. [PubMed] [Google Scholar]
- 22.Gioannini TL, et al. Endotoxin-binding proteins modulate the susceptibility of bacterial endotoxin to deacylation by acyloxyacyl hydrolase. J Biol Chem. 2007;282:7877–7884. doi: 10.1074/jbc.M605031200. [DOI] [PubMed] [Google Scholar]
- 23.Ojogun N, et al. Overproduction of acyloxyacyl hydrolase by macrophages and dendritic cells prevents prolonged reactions to bacterial lipopolysaccharide in vivo. J Infect Dis. 2009;200:1685–1693. doi: 10.1086/646616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munford RS, Sheppard PO, O’Hara PJ. Saposin-like proteins (SAPLIP) carry out diverse functions on a common backbone structure. J Lipid Res. 1995;36:1653–1663. [PubMed] [Google Scholar]
- 25.Olmeda B, García-Álvarez B, Pérez-Gil J. Structure-function correlations of pulmonary surfactant protein SP-B and the saposin-like family of proteins. Eur Biophys J. 2013;42:209–222. doi: 10.1007/s00249-012-0858-9. [DOI] [PubMed] [Google Scholar]
- 26.Hill CH, et al. The mechanism of glycosphingolipid degradation revealed by a GALC-SapA complex structure. Nat Commun. 2018;9:151. doi: 10.1038/s41467-017-02361-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorelik A, Illes K, Heinz LX, Superti-Furga G, Nagar B. Crystal structure of mammalian acid sphingomyelinase. Nat Commun. 2016;7:12196. doi: 10.1038/ncomms12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong ZJ, Huang J, Poda G, Pomès R, Privé GG. Structure of human acid sphingomyelinase reveals the role of the saposin domain in activating substrate hydrolysis. J Mol Biol. 2016;428:3026–3042. doi: 10.1016/j.jmb.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 29.Zhou YF, et al. Human acid sphingomyelinase structures provide insight to molecular basis of Niemann-Pick disease. Nat Commun. 2016;7:13082. doi: 10.1038/ncomms13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Güther ML, Leal S, Morrice NA, Cross GA, Ferguson MA. Purification, cloning and characterization of a GPI inositol deacylase from Trypanosoma brucei. EMBO J. 2001;20:4923–4934. doi: 10.1093/emboj/20.17.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McPhalen CA, Strynadka NC, James MN. Calcium-binding sites in proteins: A structural perspective. Adv Protein Chem. 1991;42:77–144. doi: 10.1016/s0065-3233(08)60535-5. [DOI] [PubMed] [Google Scholar]
- 32.Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF. GDSL family of serine esterases/lipases. Prog Lipid Res. 2004;43:534–552. doi: 10.1016/j.plipres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Kang JY, et al. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity. 2009;31:873–884. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 34.Barnes KC, et al. Polymorphisms in the novel gene acyloxyacyl hydroxylase (AOAH) are associated with asthma and associated phenotypes. J Allergy Clin Immunol. 2006;118:70–77. doi: 10.1016/j.jaci.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. Polymorphisms in RYBP and AOAH genes are associated with chronic rhinosinusitis in a Chinese population: A replication study. PLoS One. 2012;7:e39247. doi: 10.1371/journal.pone.0039247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomsen MC, Nielsen M. Seq2Logo: A method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 2012;40:W281–W287. doi: 10.1093/nar/gks469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nielsen SS, et al. BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J Appl Cryst. 2009;42:959–964. [Google Scholar]
- 45.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36:1277–1282. [Google Scholar]
- 46.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25:495–503. [Google Scholar]
- 47.Petoukhov MV, et al. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Cryst. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Cryst. 2001;34:33–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors were deposited into the Protein Data Bank (PDB) under the ID codes 5W78, 5W7C, 5W7A, 5W7B, 5W7D, 5W7E, and 5W7F.